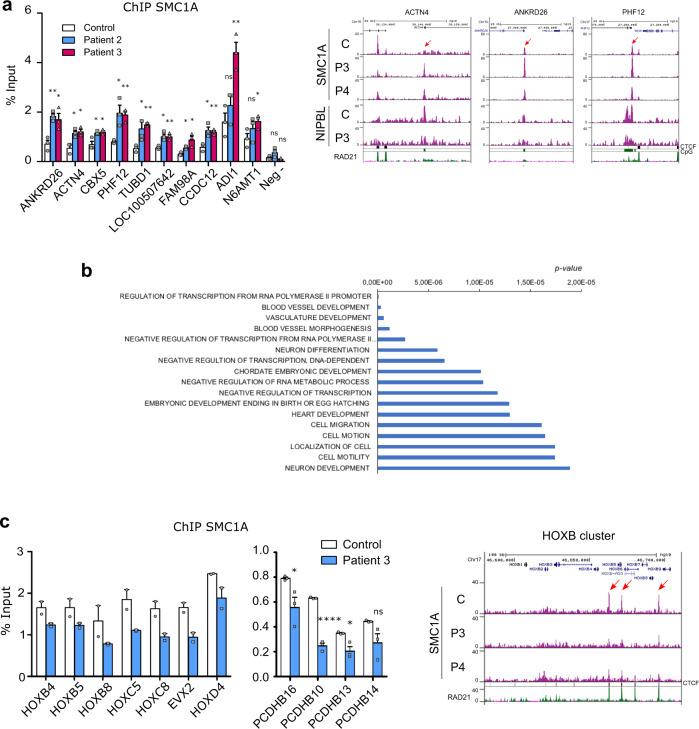
Fig. 6. Validation of the differential SMC1A peaks by ChIP-qPCR experiments.
a Cells from a control (white) and two CdLS patients (P2, blue; P3 purple) were used in ChIP-qPCR experiments to validate the SMC1A ChIP-Seq data using primers at the indicated genes. An intergenic region was used as a negative control (Neg −). Means and SEMs were calculated from biological triplicates (left). Two-sided unpaired student’s t-test (**p < 0.01; *p < 0.05). Snapshots of SMC1A and NIPBL peak distributions in control (C) and CdLS patient cells (P3–4) at three validated genome regions (right). The red arrows indicate the validated SMC1A peaks. b Gene ontology (GO) analysis reveals enriched biological processes in the SMC1A differential genomic positions in CdLS-derived fibroblasts. c ChIP-qPCR experiments were performed to validate the SMC1A ChIP-Seq data at three regions of HOXB, two of HOXC, two of HOXD, and four regions of the PCDHB clusters using cells from a control (white) and a CdLS patient (blue). Means and SEMs were calculated from biological duplicates (HOX) and triplicates (PCDHB). Two-sided unpaired student’s t-test (****p < 0.0001; *p < 0.05) (left). Snapshots of SMC1A peak distributions in control (C) and CdLS patient cells (P3–4) for the HOXB cluster (right). Red arrows indicate three validated SMC1A peaks within the HOXB cluster.