Fig. 3. The U1 snRNA-5′SS-pol II interactionlocated in the middle of an intron is necessary for splicing.
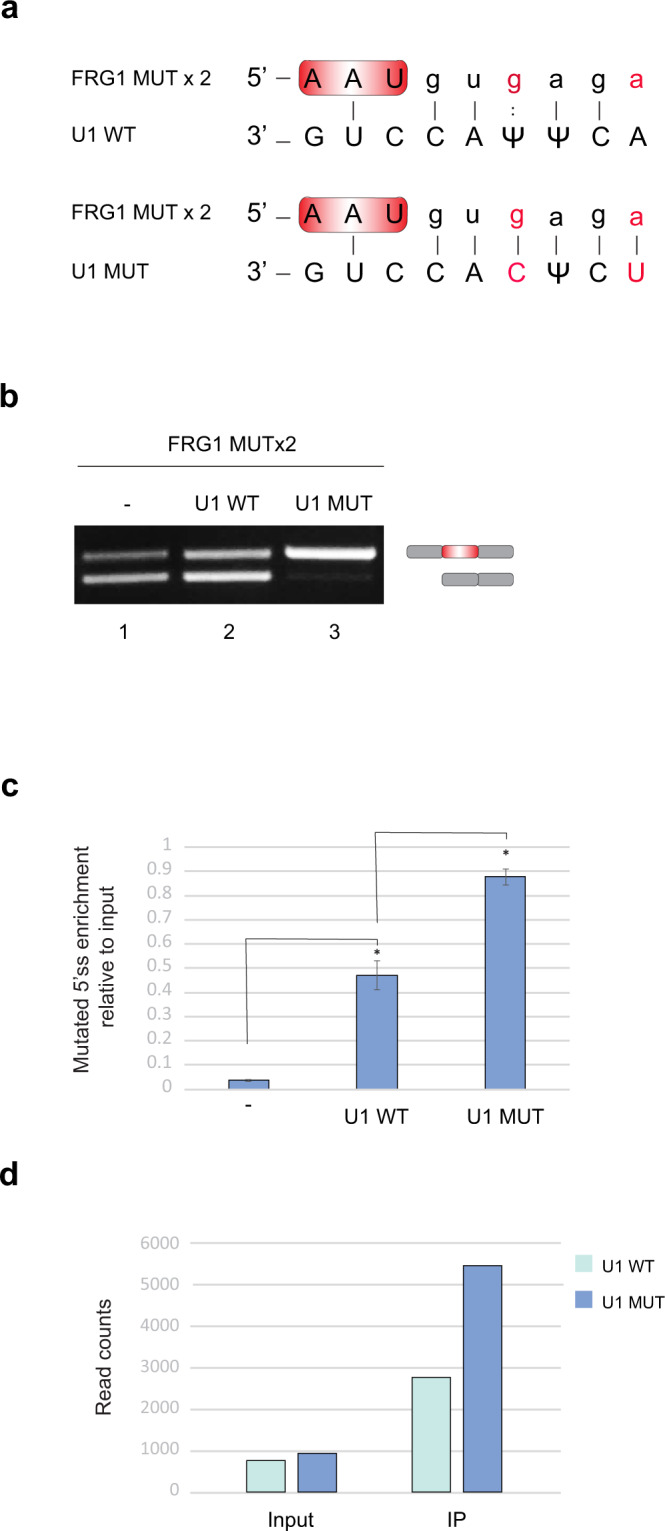
a Base pairing interaction between the mutant FRG1 (A to G and T to A at positions +3 and +6 in the second 5′SS, termed FRG1 MUTx2) and WT and MUT U1. A full line and colon indicate canonical and non-canonical base-pairing interactions, respectively. b Lane 1: RT-PCR analysis with specific primers to exons 1 and 3 of RNA extracted from cells transfected FRG1 MUTx2 minigene. Lanes 2 and 3: RT-PCR analysis of RNA extracted from cells co-transfected with FRG1 MUTx2 minigene and with either WT U1 snRNA or MUT U1, respectively. The splicing products were analyzed by gel electrophoresis. Source data are provided as a Source Data file. c The CRISPR interference protocol was performed to quantify the amount of the 5′SS of intron 2 of the FRG1 MUTx2 located in the middle of intron 2 without or with co-transfection of WT or MUT U1 snRNA. Plotted are means of n = 3 independent experiments. Error bars show mean values ± SEM. Asterisk indicates from left to right P = 0.002, 0.004, two-tailed t-test. d CRISPR interference protocol was performed after co-transfection of FRG1 MUTx2 and mutated U1 snRNA with compensatory mutations. The eluted RNA was sequenced and reads were mapped to U1 MUT and WT reference sequences. The bar plot shows the read counts in positions +3 to +6 of the U1 MUT and WT. χ2 test of independence was applied (P = 1.92e−18). One replicate was done.
