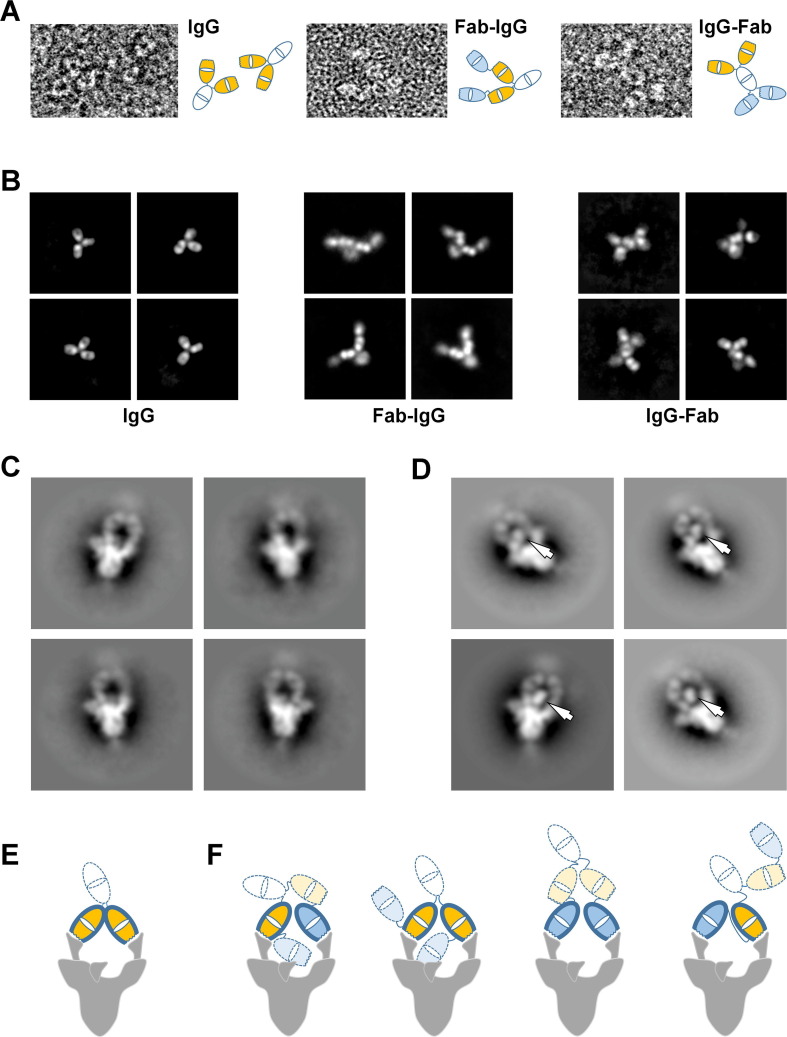
Figure 5.
Negative stain electron microscopy analysis of nAbs and their interaction with the S-protein trimer. (A) Examples of the 15033-7 IgG (left), Fab-IgG (middle) and IgG-Fab (right) molecules observed in the negative stain micrographs. Schematic interpretations are shown to the right of each image. (B) Selected negative stain electron microscopy 2D class averages for the 15033-7 IgG (left), Fab-IgG (middle) and IgG-Fab (right). (C) Negative stain electron microscopy 2D class averages of IgG 15033-7 in complex with the S-protein trimer. (D) Negative stain electron microscopy 2D class averages of Fab-IgG 15033-7 in complex with the S-protein trimer. The arrows indicate Fab-sized densities not observed in the IgG complex in (C). (E) Tentative schematic of the complex in (C) showing a trimer with 2 RBDs “up” and one RBD “down”. (F) Schematics of the four possible ways that the Fab-IgG can bind to the two “up” RBDs of a trimer that has two RBDs “up” and one RBD “down”. In (E) and (F), the following color scheme was used: white, Fc; orange, the Fabs of a native IgG; blue, the Fabs added to the native IgG. Wavy lines indicate the locations of the CDRs. The Fabs with heavy outline are making specific interactions through their CDRs. As shown by our Cryo-EM structures, the RBD in the “down” conformation is accessible for specific interaction with one of the remaining Fabs (orange or blue with light dotted outline) shown in (F). This would lead to a trivalent interaction with the S-protein trimer and may explain the additional Fab-sized densities observed in (D).