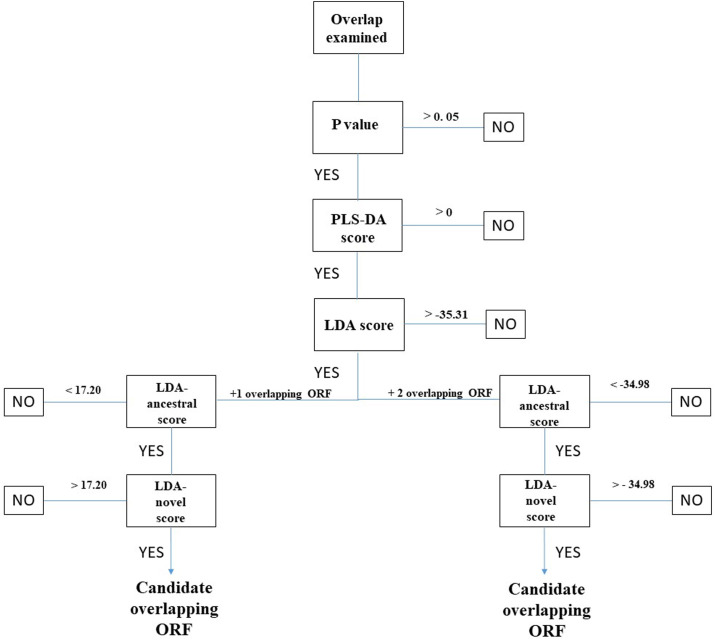
Fig. 2.
Example workflow for CodScr + SeqComp analysis. As input data, CodScr + SeqComp requires the nucleotide sequence of a protein coding region (the ancestral reading frame) which contains an overlapping ORF shifted one nucleotide 3’ (+1 overlapping ORF) or an overlapping ORF shifted two nucleotides 3’ (+2 overlapping ORF). ORF indicates a contiguous stretch of codons, beginning with a start AUG codon, ending with a stop codon, not interrupted by premature stop codons, and having a length ≥ 90 nt. A detailed example of calculation of the five prediction scores (P-value, PLS-DA score, LDA score, LDA-ancestral score, and LDA-novel score) is shown in Supplementary File S1.