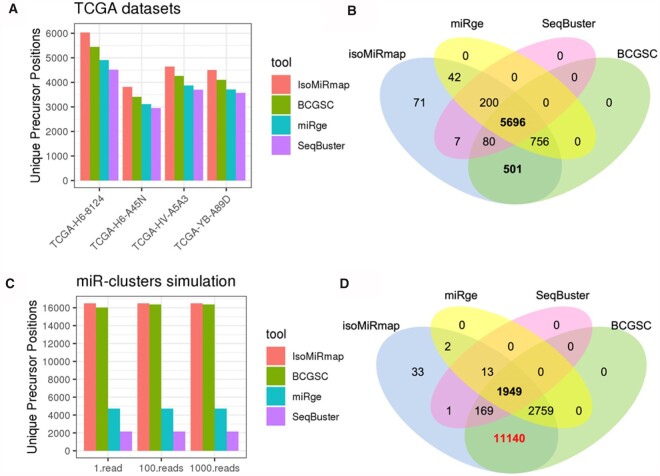
Fig. 5.
Tool comparisons. (A) Number of distinct precursor positions that can be the source of wild-type isomiRs that are reported by each of the studied tools. The analyzed datasets comprised four datasets from the TCGA repository (see text). (B) Overlap of the unique wild-type isomiR sequences that were reported by each tool for the TCGA datasets. (C and D) Similar to A and B respectively but for the three synthetic datasets comprising all possible wild-type isomiRs from the let-7 family, mir-17/92 (plus paralogues) cluster and miR-320 family (see text)