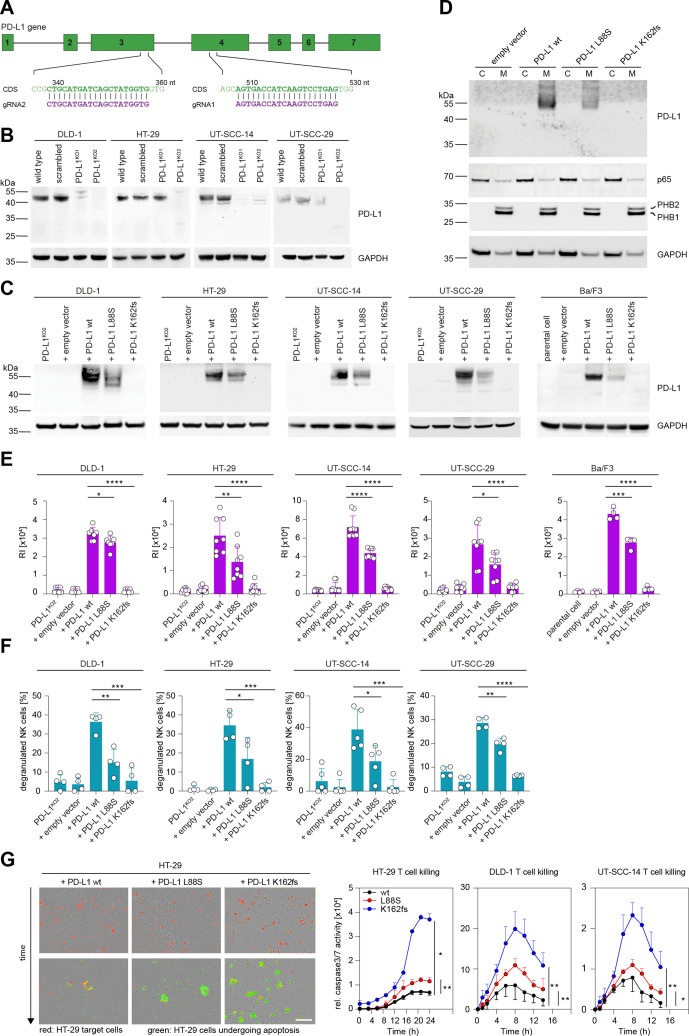
Figure 5.
Selected PD-L1 mutations reduce protein abundancy, surface expression, ADCC and T cell suppression. (A) Schematic representation of gRNAs targeting the PD-L1 coding sequence (CDS). (B) CRISPR/Cas9-mediated depletion of PD-L1 as detected in crude cell extracts via immunoblotting. (C) PD-L1 protein levels after lentiviral redelivery of PD-L1 variants into PD-L1KO2 human cell lines and the murine Ba/F3 cell line as detected by immunoblotting. (D) Enrichment of PD-L1 in the membranous fraction (M) of HT-29 cell variants from panel C. (C) Cytosolic, non-membranous fraction. (E) Flow cytometric detection of PD-L1 surface expression displayed as mean relative fluorescence intensity (RI) after staining with avelumab. n(DLD-1)=7, n(HT-29)=8, n(UT-SCC-14)=9, n(UT-SCC-29)=8, n(Ba/F3)=4. (F) NK cell degranulation induced by coculturing primary NK cells and cell lines expressing indicated PD-L1 variants in the presence of avelumab (n=4 for DLD-1, HT-29, UT-SCC-29; n=5 for UT-SCC-14). Percent degranulated NK cells normalized to spontaneous NK degranulation is shown for all cocultures. (G) Representative images and time course of T cell mediated tumor cell killing. CD8+ T cells were cocultured with HT-29, DLD-1 and UT-SCC-14 cells expressing WT PD-L1 or the L88S and K162fs variants (red fluorescence). Caspase 3/7 activity (relative intensity of green fluorescence) was monitored every 90 min for 24 hours with an Incuyte S3. Scale bar, 200 µM. Asterisks indicate p value range (*p<0.05; **p<0.01; ***p<0.001; ***p<0.0001). Statistics for time course: two-tailed paired t test. All other statistics: two-tailed unpaired t test. NK, natural killer; PD-L1, programmed cell death protein ligand 1.