Figure 1.
Paired Cas9-D10A nickases induce efficient genome editing of PINK1 in cynomolgus monkey embryos
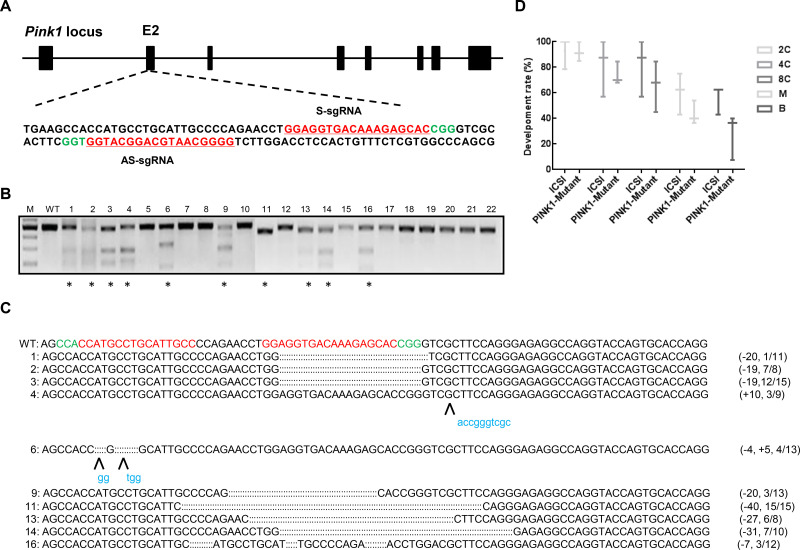
A: Schematic of sgRNAs targeting PINK1 loci. Guide RNA sequences are underlined and highlighted in red. PAM sequences are highlighted in green. B: Cas9-mediated on-target cleavage of PINK1 by T7E1 cleavage assay. PCR products were amplified and subjected to T7E1 digestion. Samples with cleavage bands are marked with an asterisk. M, marker; WT, wild-type. C: Editing profiles of marked samples in (B). Undigested PCR products from (B) were subjected to TA cloning. Single TA clones were selected and analyzed by DNA sequencing. For WT alleles, PAM sequences are highlighted in green and sgRNA sequences are labeled in red. For alleles with indels, deleted bases are replaced with colons and inserted bases are labelled in lower case and highlighted in blue; deletions (-) and insertions (+). D: Developmental rate in Cas9-D10A-injected embryos is comparable to that in ICSI-treated embryos. ICSI, intracytoplasmic sperm injection embryo; PINK1-Mutant, PINK1 sgRNA injected embryo; 2C, 2-cell embryo; 4C, 4-cell embryo; 8C, 8-cell embryo; M, morula; B, blastula.