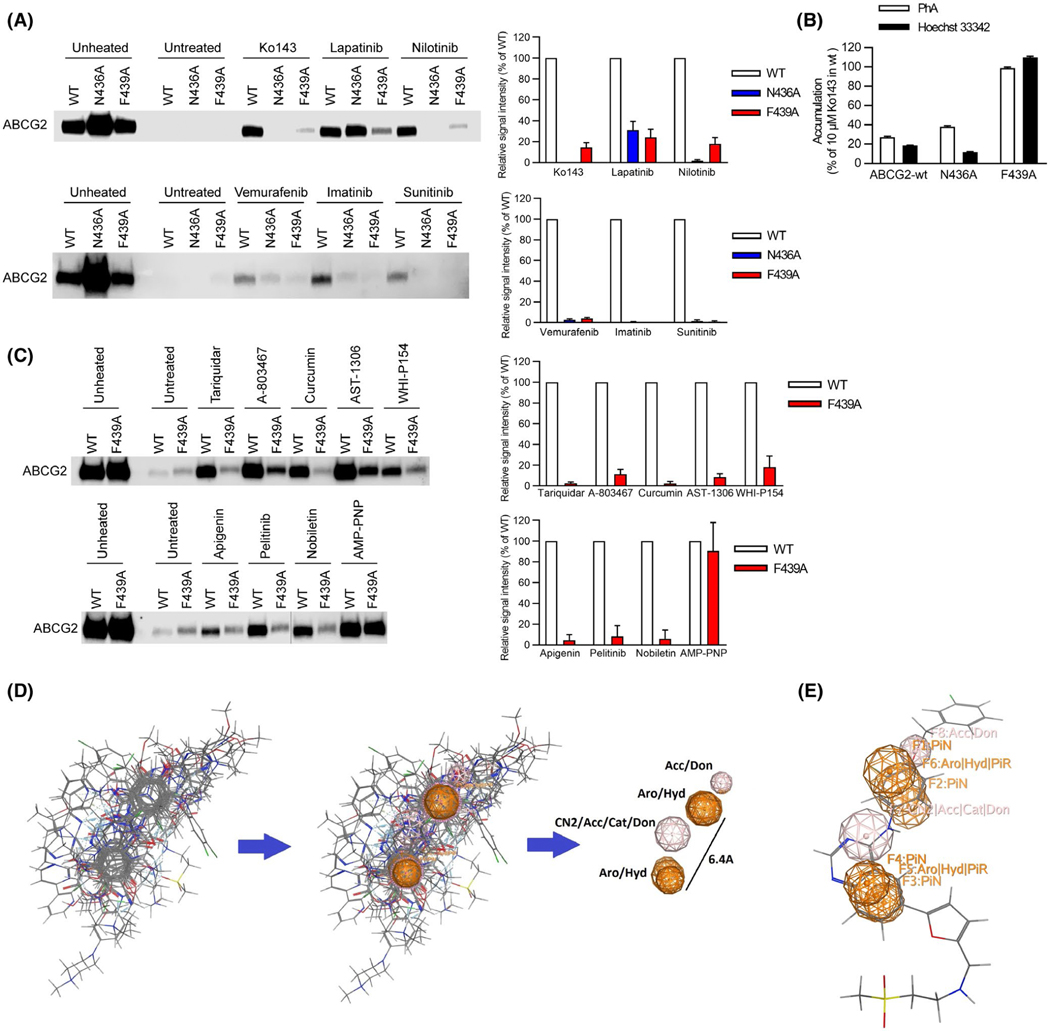
FIGURE 3.
A key amino acid residue in the binding pocket is required for binding to ABCG2. A, Role of N436 and F439 in kinase inhibitor binding assessed by thermal shift. Representative western blots are shown on the left side and the densitometry quantification data from three to four independent experiments are shown on the right side. Membrane vesicles prepared from mAbcg2-KO MEFs expressing human ABCG2 (WT, N436A, and F439A) were incubated with 10 μM of kinase inhibitor for 60 min at 37°C. Samples were heated for 3 min at 65°C for WT-ABCG2 (white), 67°C for N436A (blue), and 61°C for F439A mutant (red). B, N436A retains transport of Hoechst 33342 (black) and PhA (white), whereas F439A has lost transport ability. Each bar represents the mean + SE (n = 8). C, F439 contribution to binding of various ligands to ABCG2. Representative western blots are shown on the left side and the densitometry quantification data from three independent experiments are shown on the right side. Membrane vesicles prepared from mAbcg2-KO MEFs expressing human ABCG2 (WT and F439A) were incubated with 10 μM of ABCG2 ligand for 60 min or 1 mM of AMP-PNP for 30 min at 37°C. Samples were heated for 3 min at 65°C for WT-ABCG2 (white) and 61°C for F439A mutant (red). D, Best flexible alignment of all ABCG2 ligands after a 48 hr alignment run in MOE, was used to compute the consensus pharmacophore model that had two aromatic features at 6.4Å to each other. E, Docked Lapatinib molecule mapped onto the consensus pharmacophore model, showing the connection between the docking model and the consensus pharmacophore model. Don = H-bond donor; Acc = H-bond acceptor; PiN = Ring normal; Aro = Aromatic; PiR = pi-Ring; Hyd = Hydrophobic; Cat = Cation (use MOE reference)