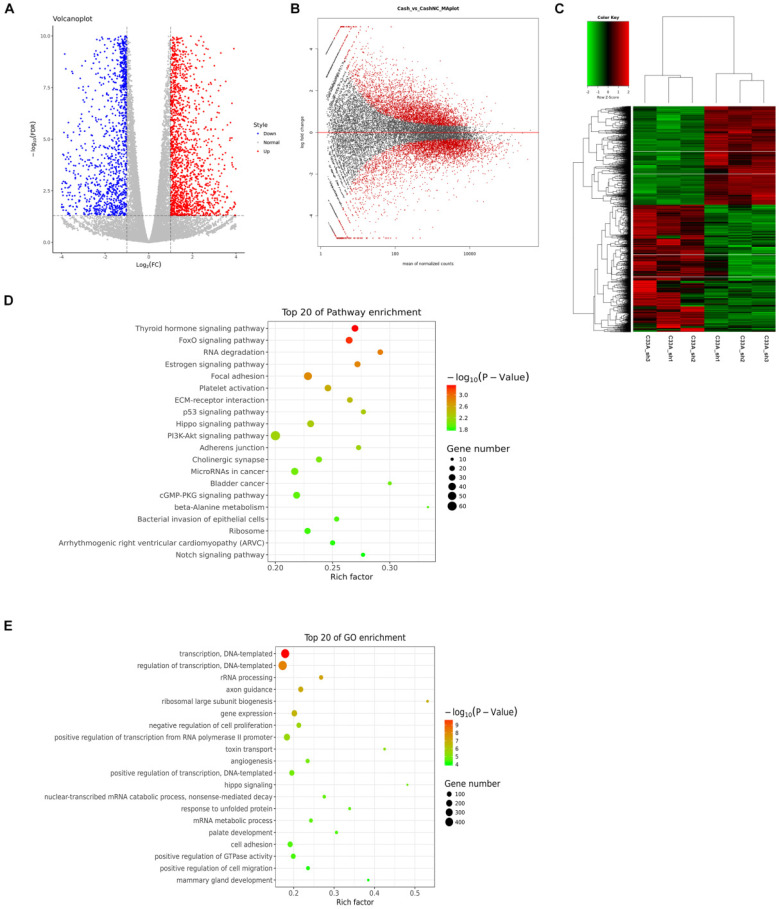
Figure 4.
Analyses of differential genes in cyclin D1 knock-down C33A cells. (A and B) Each dot represents a gene, the red dots represent up-regulated differentially expressed genes, the blue dots in the figure represent down-regulated differentially expressed genes, and the gray dots represent non-differentiated genes. While the red dots represent differentially expressed genes, and the black dots represent non-differentiated genes. (C) The heat maps displayed genes differentially regulated by cyclin D1, the three biological replicates also had good consistency from the hierarchical clustering analyses. (D) The differentially expressed genes with KEGG (Kyoto Encyclopedia of Genes and Genomes: systematic analysis of gene function or genomic information database). We selected the top 20 of pathway enrichment which were most significant based on the Rich factor and p value, it was found that these genes involved several important pathways such as PI3K-Akt signaling pathway. (E) The top 20 of pathway enrichment by GO-analysis (Gene Ontology), it was found that these genes involved several important pathways such as transcription.