Figure 1.
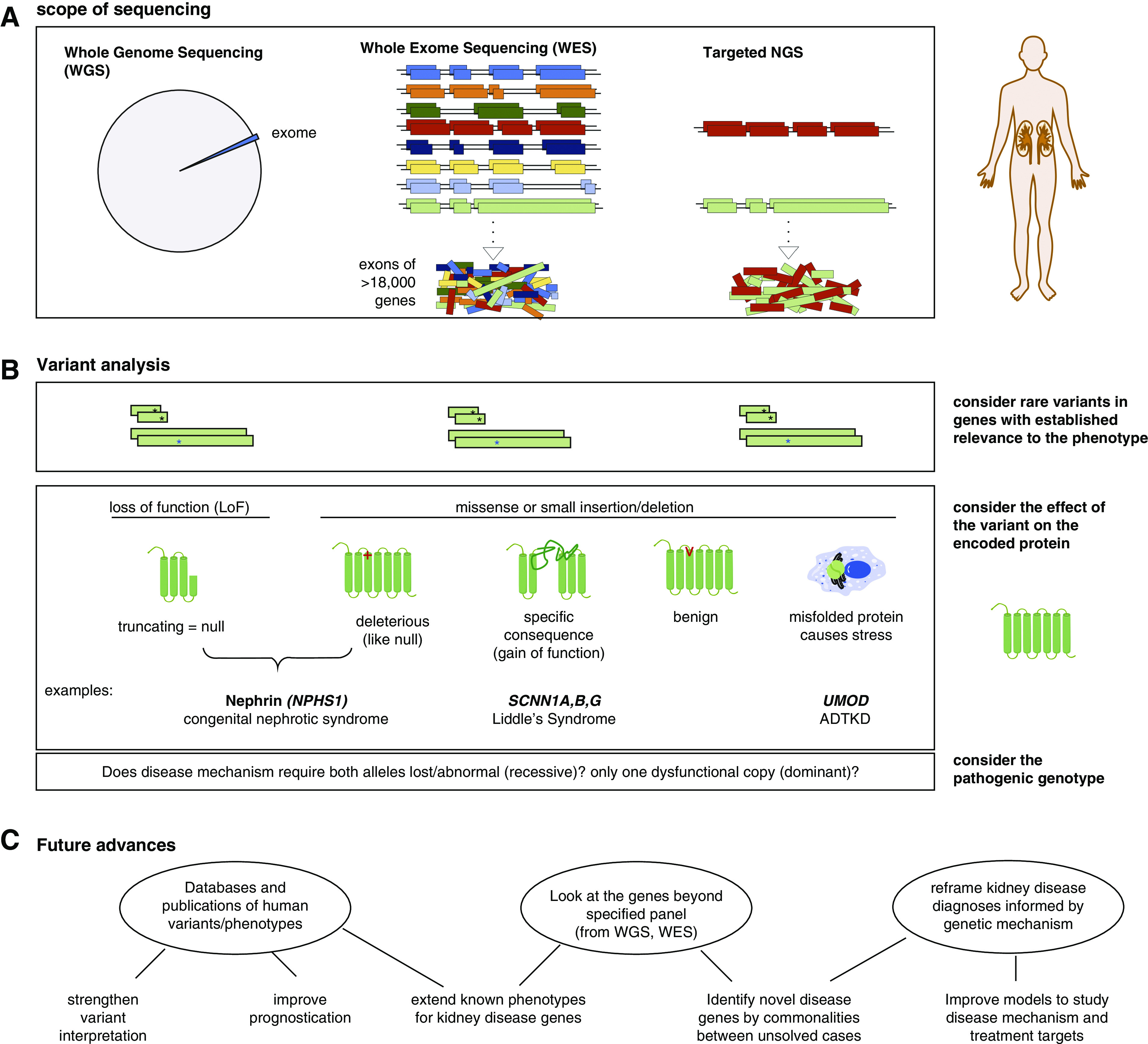
Next generation sequencing (NGS) applied to kidney disease analysis. (A) NGS can be applied to the whole genome (whole-genome sequencing [WGS]), the exome (whole-exome sequencing [WES]), or specific genes (targeted NGS). (B) Variant analysis of a gene panel proceeds similarly regardless of scope of the sequencing. A “pathogenic” variant must be more rare in the general population than the rare kidney phenotype and either encode a premature truncation of the protein or have an established causative association with disease in public databases. Other rare nontruncating variants can be considered “likely pathogenic” on the basis of frequency and predictive algorithms that consider whether the affected amino acids have remained unchanged (“conserved”) through species evolution and are in critical protein domains or other regulatory features. Beyond the consequence of the variant on the protein, interpretation of its effect must include knowledge of the disease mechanism and whether monoallelic (heterozygous) or biallelelic (homozygous or compound heterozygous) mutation is needed for pathogenesis. ADTKD, autosomal dominant tubulointerstitial kidney disease. (C) Applying genetic analysis to kidney disease evaluation has several potential future benefits.
