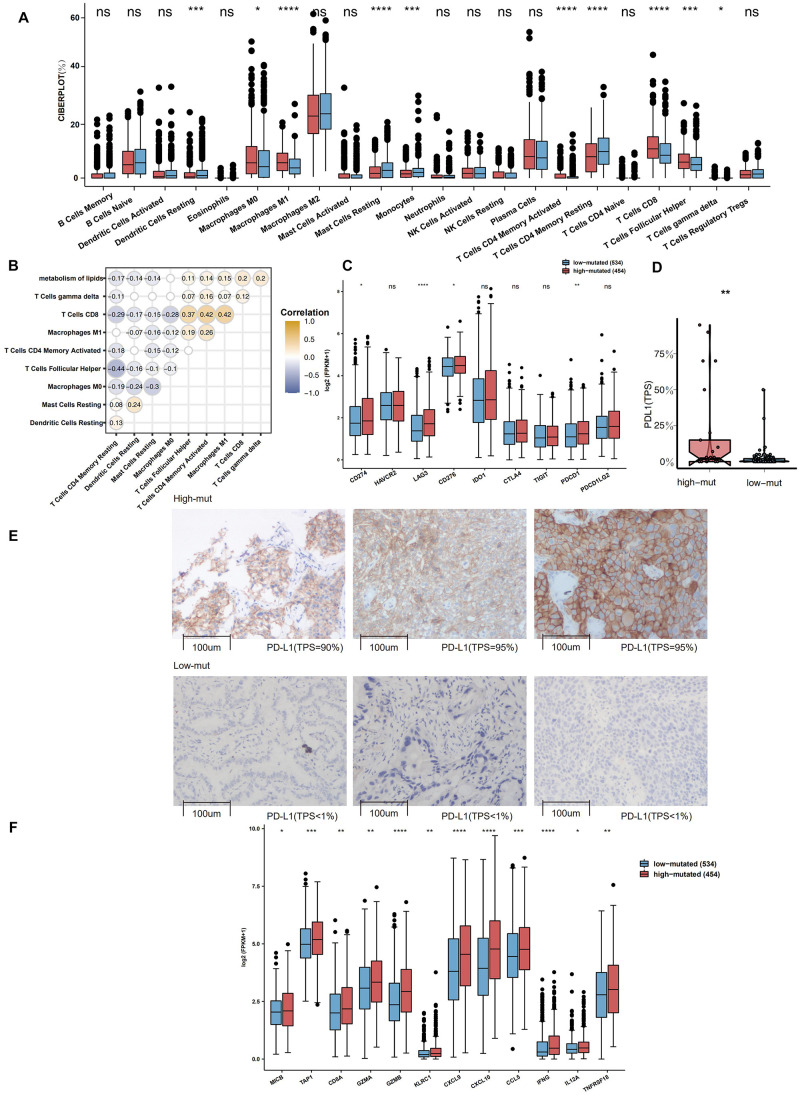
FIGURE 3.
(A) Comparison of the fractions of 22 types of immune cells as estimated by the CIBERSORT algorithm between the high and low mutation groups in the TCGA cohort. (B) The correlations between the number of mutations in the lipid metabolism pathway and the proportions of immune cells. (C) Comparison of the expression of immune-related genes between the high and low mutation groups in the TCGA cohort. (D) Comparison of the expression of PD-L1 (TPS) between the high and low mutation groups in the Local-NSCLC cohort. (E) The typical cases for each TPS level between the high (three samples; high PD-L1 TPS) and low mutation (three samples; no PD-L1 TPS) groups in the Local-NSCLC. Using HE and PD-L1 stained slides, we manually assessed the number of tumor cells, the sample size (diameter), the crush rate with a cut-off value of <1% (no PD-L1 TPS), 1–50% (low PD-L1 TPS), 50% < (high PD-L1 TPS), and the TPS for each biopsy sample using the slide that contained the most tumor cells. The TPS level was evaluated by pathologists who completed training courses in TPS estimation. (F) Heatmap depicting the mean differences in the expression of proinflammatory and antigen presentation genes between the high and low mutation groups in the TCGA cohort. Each square represents the fold change or the mean difference in the expression of these genes between the high and low mutation groups in the TCGA cohort. Red indicates upregulation.