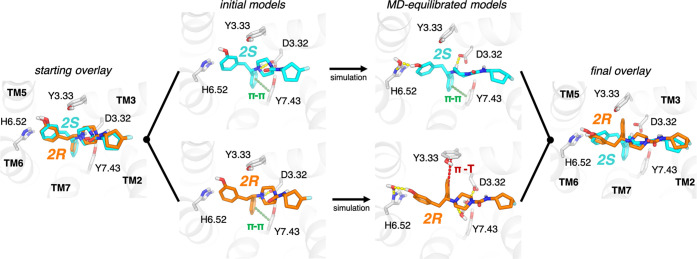
Figure 2.
MD simulations reveal distinct poses for different ligand enantiomers. Simulations of pairs of enantiomers (e.g., 2S and 2R shown) were initiated from docked poses. The S- and R-enantiomers formed similar interactions in their initial poses (left), including a π-stack with Y7.43 and a salt bridge with D3.32. During simulations, 2S maintained all of the initial interactions while also forming a water-mediated interaction with H6.52 (top right). By contrast, 2R underwent a reorientation, breaking the initial π-stack with Y7.43 and instead reorienting toward Y3.33 in a potential T−π stack interaction, while it retained the water-mediated interaction with H6.52 (bottom right). This reorientation also significantly weakens the direct salt bridge interaction with D3.32 for 2R. An overlay of the final poses (right) highlights the difference in the orientation of the phenyl group attached to the chiral center.