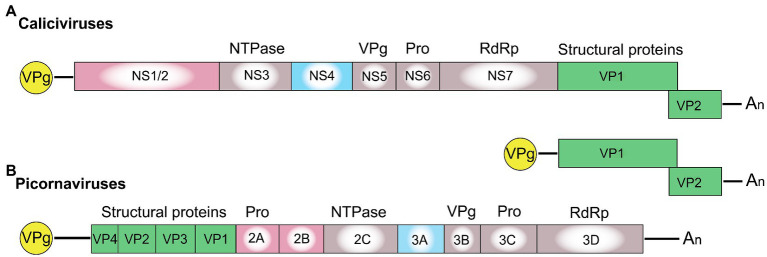
Figure 1.
Schematic representation of the genome organization for a typical (A) calicivirus and (B) picornavirus. Untranslated and translated sequences are depicted as lines and boxes, respectively. Structural (capsid) proteins are shown in green (note that VP1 and VP2 in lagoviruses are usually referred to as VP60 and VP10, respectively); the signature proteins of the picornavirus-like superfamily are shown in gray, namely the helicase (NTPase), VPg, protease (Pro), and the RNA-dependent RNA polymerase (RdRp); the non-structural calicivirus proteins NS1/2 and their positional homologs in picornaviruses (2A, 2B) are shown in pink; and the non-structural calicivirus protein NS4 and its homolog in picornaviruses (3A) are shown in blue. Covalently linked VPg proteins at the 5' end are shown in yellow, and An represents the poly(A) tail at the 3' end.