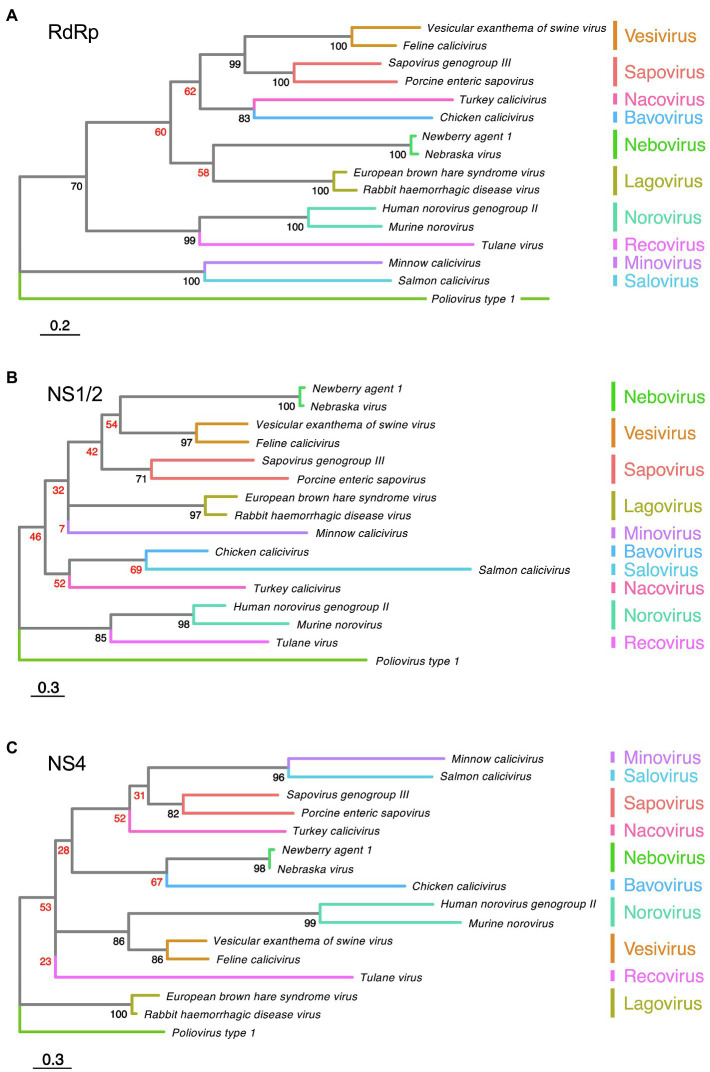
Figure 2.
Phylogenetic analysis of calicivirus protein sequences. Maximum likelihood phylogenies were inferred for amino acid sequences of (A) RdRp, (B) NS1/2, and (C) NS4. First, the amino acid sequences of the complete ORF1 coding region of representative published calicivirus sequences were aligned using MAFFT (Katoh et al., 2002). The alignment was then curated with trimAl (Capella-Gutiérrez et al., 2009). The RdRp, NS1/2, and NS4 coding regions were extracted from this complete ORF1 alignment and phylogenies were inferred individually for each gene using IQ-TREE (Nguyen et al., 2015). Phylogenies were rooted using Poliovirus type 1 (GenBank accession NC_002058). The following sequences were chosen for calicivirus genera: Vesicular exanthema of swine virus (VESV; NC_002551), Feline calicivirus (FCV; NC_001481), Sapovirus genogroup III (MG012434), Porcine enteric sapovirus (NC_000940), Turkey calicivirus (NC_043516), Chicken calicivirus (NC_033081), Newbury agent 1 (NC_007916), Nebraska virus (NC_004064), Rabbit haemorrhagic disease virus (NC_001543), European brown hare syndrome virus (EBHASV; NC_002615), Tulane virus (NC_043512), Human norovirus genogroup II (NC_039477), Murine norovirus (MNV; NC_008311), Minnow calicivirus (NC_035675), and Salmon calicivirus (NC_024031). The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. Ultrafast bootstrap values are shown for each node. Low confidence bootstrap values (<70) are highlighted in red.