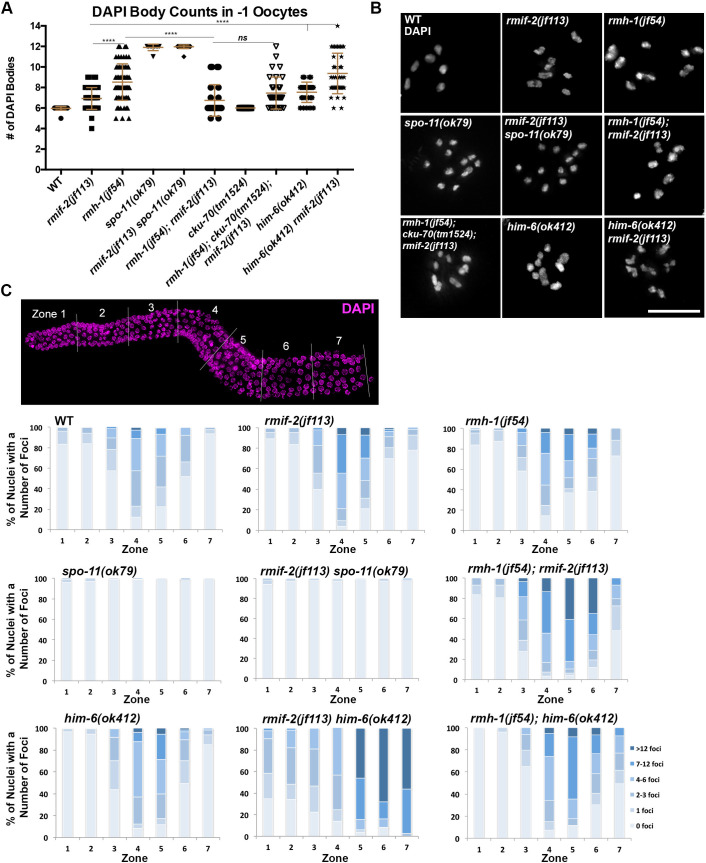
Fig 2. RMIF-2 is required for robust chiasma formation and chromosome segregation in meiosis.
(A) Quantification of DAPI-stained bodies in -1 diakinesis oocytes in the WT (number of nuclei, n = 32), rmif-2(jf113) (n = 41), rmh-1(jf54) (n = 74), spo-11(ok79) (n = 12), rmif-2(jf113) spo-11(ok79) (n = 32), rmh-1(jf54); rmif-2(jf113) (n = 27), cku-70(tm1524) (n = 19), rmh-1(jf54); cku-70(tm1524); rmif-2(jf113) (n = 33), him-6(ok412) (n = 26), and rmif-2(jf113) him-6(ok412) (n = 41) mutants. Data are the mean and standard deviation (error bars). Significant differences were determined using a Student T-test: **** p < 0.0001. (B) Representative images of chromosomes in a diakinesis nucleus for each genotype stained with DAPI. Scale bar: 10μm. (C) Quantification of RAD-51 profiles throughout meiotic prophase I (upper panel). C. elegans gonads were divided into seven equal zones. RAD-51 foci were counted in each nucleus of each zone; three representative gonads per genotype. Graphs show the percentage of nuclei with different numbers of foci per germline zone. Raw data and statistical analysis of RAD-51 profiles between different zones and genotypes via Fisher’s exact test are presented in S2 File. Scale bar: 10μm.