Figure 6: Engraftment modeling is accurate in a meta-analysis of five FMT trials for the treatment of recurrent C. difficile.
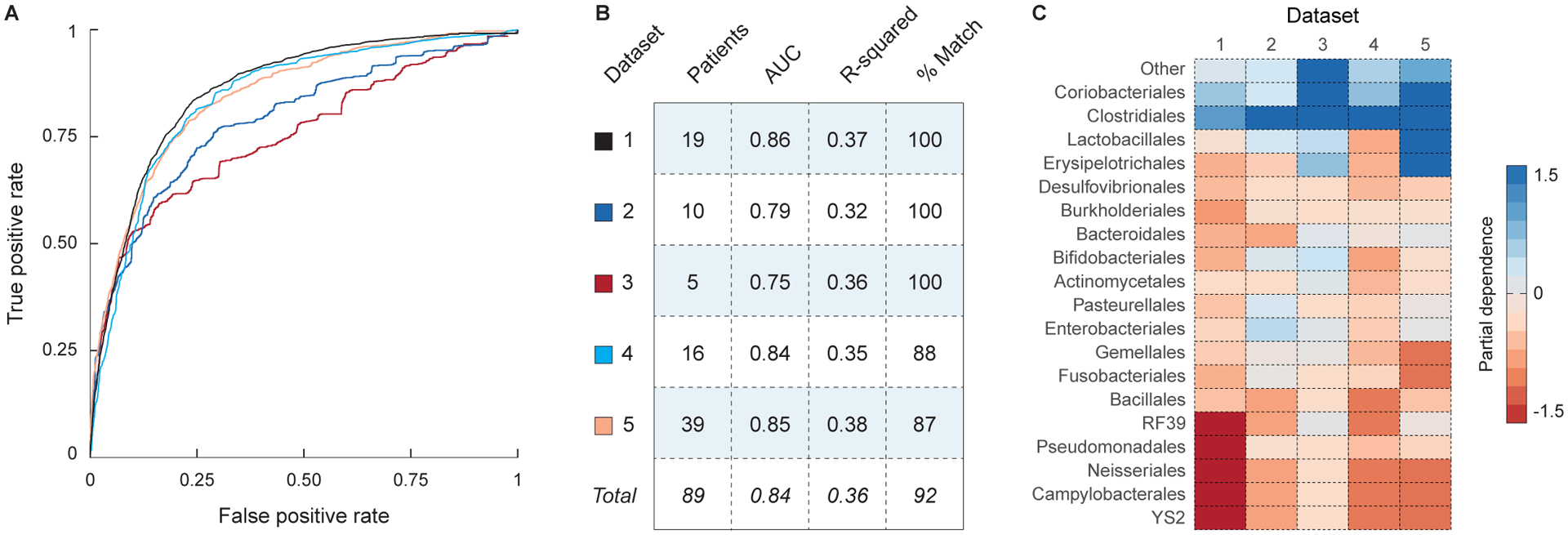
Models of bacterial engraftment were trained on the 16S rRNA sequence data from five FMT trials for the treatment of recurrent C. difficile infection. (A) ROC curves for the predictions of OTU presence in each of the five FMT datasets (see legend in panel B). (B) Statistics describing the performance of each model, including the number of patients in the dataset, the AUC (for predictions of OTU presence), the r-squared (for predictions of OTU abundance), and the percent of predictions that correctly cluster with their target samples (see Methods). (C) Heatmap showing the partial dependence of the models of OTU presence on each bacterial order. High values indicate that the taxon has a favorable impact on engraftment.
