Fig. 1. HIV Env protein in nsEM.
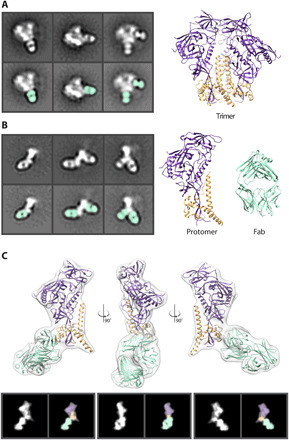
(A) Negative stain 2D classes of Env trimer with Fabs bound. Fabs are highlighted in seafoam green. Cryo–electron microscopy (cryo-EM)–derived model of Env BG505 SOSIP trimer (PDB: 6DID). gp120 (purple) and gp41 (orange). (B) Negative stain 2D class of Env protomers with Fabs bound. Models of a single Env protomer and Fab [Protein Data Bank (PDB): 6DID] in seafoam green are shown as high-resolution models for comparison. (C) Low-pass–filtered cryo-EM map of an Env protomer bound to 1C2 Fab (PDB: 6P65) were used to generate 2D back projections, which clearly reveal the gp120 region as a wider density at the apex. This easily identifiable topology in the 2D classes enables assignment of the orientation of protomer and subsequent identification of epitopes to be determined.
