Fig. 3. The m6A modification expedites the degradation of HMBOX1 mRNA per se, which is mediated by the reader protein YTHDF2.
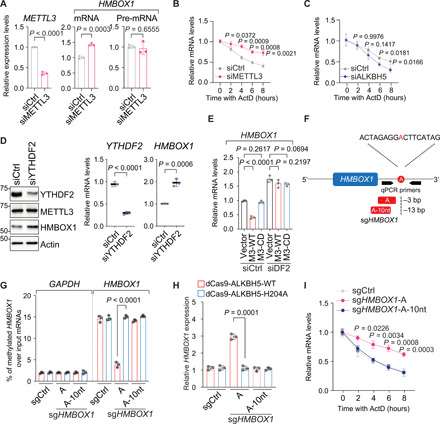
(A) Levels of specified mRNA molecules in LNCaP cells transfected with control siRNA (siCtrl) or siRNA targeting METTL3 (siM3) for 72 hours. (B and C) Half-life of HMBOX1 mRNA in LNCaP cells upon silencing METTL3 (B) or ALKBH5 (C) with the treatment of ActD (5 μg/ml) before total mRNAs were collected at indicated time points. (D) Levels of indicated molecules upon YTHDF2 knockdown in LNCaP. (E) Expression of HMBOX1 in the control (siCtrl) and YTHDF2-knockdown (siDF2) cells of LNCaP stably expressing empty backbone (vector), wild-type METTL3 (M3-WT), or catalytically dead mutant (M3-CD). (F) Schematic illustration of effective (A) and noneffective (A-10nt) sgRNA targeting HMBOX1 transcript (sgHMBOX1). (G) MeRIP-qPCR analysis of m6A signals on HMBOX1 in LNCaP cells expressing dCas9 fused to the wild-type ALKBH5 (dCas9-ALKBH5-WT) or to the incompetent demethylase (dCas9-ALKBH5-H204A) together with control sgRNA (sgCtrl), sgHMBOX1-A, or sgHMBOX1-A-10nt. GAPDH was included as a negative control. (H) Expression of HMBOX1 in the CRISPR-dCas9–based, m6A-editing system that is described in (G). (I) Half-life of HMBOX1 mRNA in LNCaP cells expressing dCas9-ALKBH5-WT together with specified sgRNAs and treated with ActD as described in (B) and (C).
