Fig. 4. Long KLCCTDs bind to specific anionic phospholipids.
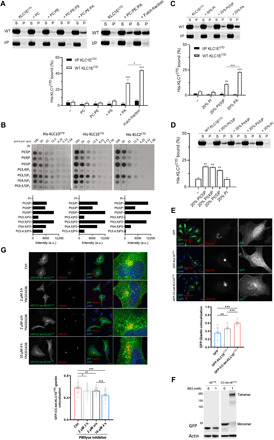
(A) Cosedimentation analysis of WT or I/P His-KLC1ECTD incubated with PC, PC:PE (7:3), PC:PE:PS (5:3:2), PC:PE:PA (5:3:2), or Folch fraction I LUV. (B) Binding of the indicated long KLCCTDs to phosphoinositides in a protein lipid overlay assay. Quantitation is provided at the 50 pmol per spot. a.u., arbitrary units. (C) Cosedimentation analysis of WT or I/P His-KLC1ECTD incubated with PC:PE:PI, PC:PE:PI(5)P, or PC:PE:PA (5:3:2) LUV. (D) Cosedimentation analysis of WT His-KLC1ECTD incubated with PC:PE:PI(3)P, PC:PE:PI(4)P, PC:PE:PI(5)P, or PC:PE:PI (5:3:2) LUV. Means ± SEM of at least three experiments. *P < 0.05, **P < 0.01, and ***P < 0.001 compared to the sample without liposomes or as indicated in the graph. (E) Immunofluorescence analysis (top) showing localization of GFP, GFP-KLC1ECTD, and GFP-CC-tet-KLC1ECTD. Golgi is stained with anti-giantin antibody. Representative Pearson’s correlation from one experiment and ≈40 cells (bottom). (F) BS3 cross-linking assay showing monomeric or tetrameric expression of KLC1ECTD in lysates of HeLa cells transfected with GFP-KLC1ECTD or GFP-CC-tet-KLC1ECTD. (G) GFP-CC-tet-KLC1ECTD-giantin colocalization in cells treated with the PIKfyve inhibitor YM201636 as indicated (top). Representative Pearson’s correlation from one experiment and ≈40 cells (bottom). Means ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001. Scale bars, 20 μm. DAPI, 4′,6-diamidino-2-phenylindole.
