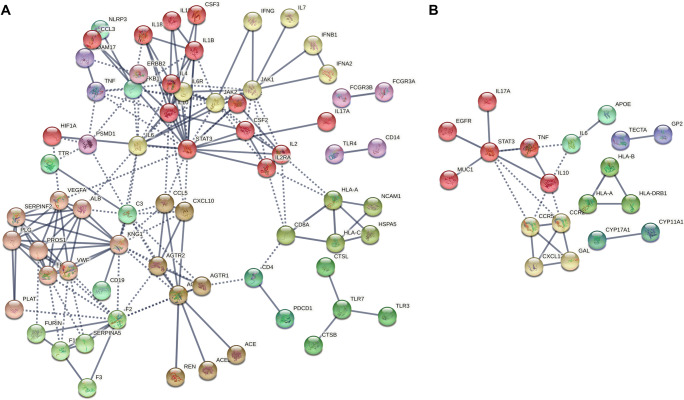
Figure 1.
Functional protein-protein correlation networks of the COVID-19 (A) and herpes zoster-related (B) genes were determined using STRING. Genes correlated with these 2 disease entities (herpes zoster, disease id: C0019360, Gene-Disease Association Score ≥ 0.01; COVID-19, id: C000657245, version 5, N.PMIDs ≥ 10) were retrieved from the DisGeNET, a platform for the disease genomics. Only highly confident interactions with minimum required interaction score of 0.900, derived from “databases,” “co-occurrence,” “co-expression,” and “experiments,” were counted. Network clustering was done with the MLC (Markov cluster) algorithm with an inflation parameter of 3