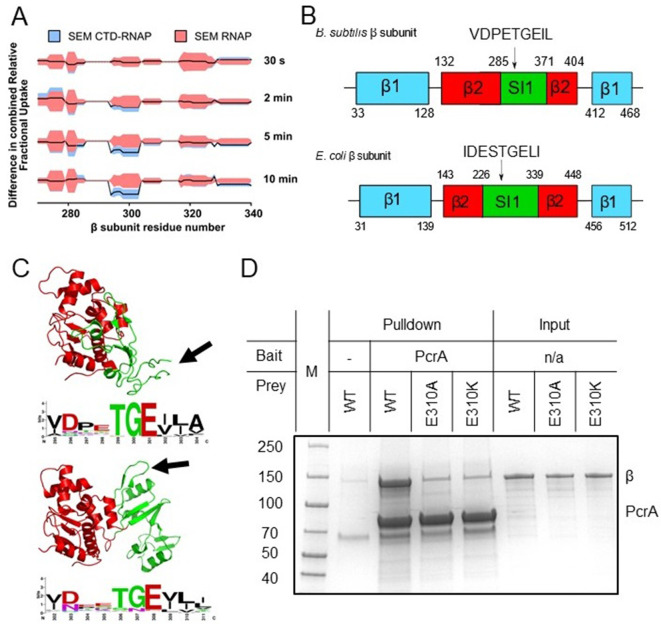
Figure 2. The PcrA-CTD binds to a conserved motif in the SI1 domain of RNAP.
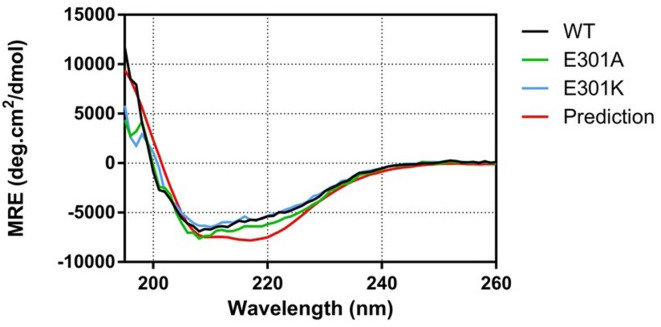
(A) Relative HDX measured for a region of the RNAP β subunit (residue numbers on x axis) within the CTD-RNAP complex (blue) compared to RNAP alone (red). A small region of RpoB (at amino-acid positions around ~300) becomes significantly protected by interaction with the PcrA CTD as the exchange time becomes longer. (B) The protected region maps to a conserved motif in the SI1 domain of B. subtilis RpoB. This region is organised differently in E. coli RpoB, but the same conserved amino acid motif appears in a slightly different position in the structure (black arrow). (C) Structure of the B.subtilis (upper panel) and E. coli (lower panel) β2 (red) - SI1(green) domains indicating the beta-loop structure containing a putative interaction motif at the tip (black arrows). This sequence is well-conserved in bacterial RNA polymerases and the consensus sequence is shown in weblogo format beneath each structure. (D) In vitro pulldown of RpoB using PcrA as a bait (see Materials and methods for details). Mutation of the conserved glutamate (E301) in the putative helicase interaction motif dramatically reduces RpoB pulldown.