Abstract
Purpose
Wilms tumor 1 (WT1) gene has recently shown a role in gliomagenesis, making it a potential immunotherapy target in glioblastomas. We aimed to investigate the most sensitive method to detect WT1 expression in glioblastoma and explore the relationship between WT1 expression, IDH1 mutation and recurrence interval.
Patients and Methods
Clinical data were collected from 44 patients with glioblastomas, treated with adjuvant therapies. WT1 expression was assessed in all cases using immunohistochemistry (IHC), while its gene expression was assessed in 13 clustered samples using polymerase chain reaction (qPCR). IDH1 mutation was assessed using IHC. The sensitivity between IHC and RT-qPCR was examined. Kaplan–Meier curves were used to compare the recurrence-free interval (RFI) between IDH1 and WT1 expression groups.
Results
IDH1wildtype was found in 26 cases (59.1%) and the remaining 18 cases (40.9%) were IDH1mutant. Through IHC, WT1 was overexpressed in 32 cases (72.7%), partially expressed in 9 cases (20.5%) and not expressed in only 3 cases. For the 13 cases tested by qPCR, 6 cases showed WT1 upregulation and 7 cases showed WT1 downregulation. There was no significant difference in WT1 expression among cases with different RNA concentrations regardless the testing method (p-value >0.05). However, the difference between IHC and qPCR was significant. IDH1mutant cases with WT1 overexpression showed significant difference in RFI (p-value =0.048).
Conclusion
Parallel testing for WT1 expression using IHC and qPCR is not reliable. However, IHC provides more accurate results. Moreover, IDH1mutant glioblastomas with WT1 overexpression are associated with late RFI particularly if temozolomide with additional chemotherapies are used.
Keywords: glioblastoma, IDH1 mutation, WT1 expression, chemotherapies, PCR sensitivity
Introduction
Wilms tumor 1 (WT1) gene encodes a zinc finger transcriptional factor that plays an important role in cell growth and differentiation.1 WT1 has been implicated in various malignancies. It was first identified as a tumor suppressor gene because of frequent chromosome 11p13 region deletions observed in childhood renal neoplasm and Wilms tumor and was then found to be overexpressed in leukemias and various solid tumors including breast and ovarian cancers2–4 Few studies have shown that WT1 has a role in gliomagenesis.5 Consistently, WT1 overexpression has been found in high‑grade gliomas.4,6,7 Recent clinical trials of cancer immunotherapy targeting WT1 protein have shown promising results in glioblastomas, particularly in resistant cases. These results suggested that WT1 is a possible target for immunotherapy in high‑grade gliomas, which can increase the sensitivity of glioblastoma to chemoradiotherapy.8
The immunohistochemical approach is considered as the standard method to detect WT1 protein expression in tumour cells. However, some studies have shown that WT1 mRNA levels present results similar to that of the immunohistochemical score.4 Therefore, the most accurate method for testing WT1 expression in glioblastoma is not obviously clear. Our study was designed to investigate whether IHC or qPCR is more sensitive for detecting WT1 gene expression in glioblastoma cases.
Rauscher et al found that some high-grade gliomas lacked WT1 expression, whereas Manocha et al identified an inverse relationship between WT1 scores and IDH1 mutation.8,9 They ascribed this negative expression in high‑grade tumors to the younger age of patients and tumors possessing IDH1 mutations.10 Our study was also designed to explore the relationship between WT1 expression and IDH1 mutation and how this influences tumour recurrence.
Patients and Methods
Sample Stratification
We included 44 patients with totally resected glioblastomas, who received adjuvant therapies, in the period between 2015 and 2019. Ethical approval for this study was granted by the National Biomedical Ethics Committee at King Abdulaziz University (HA-02-J-008) (Reference No. 189-19). All patients involved in this study have provided informed consent. All procedures performed in this study were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. Histological diagnoses were made according to the World Health Organization (WHO) classification. Clinical data including age at diagnosis, gender, postoperative adjuvant therapies, type of chemotherapies, and recurrence interval were retrieved from hospital records. Patients were stratified based on IDH1 mutation and WT1 expression (Figure 1). Standard radiotherapy of a total dose of 60 Gy and temozolomide (TMZ) (150–200 mg/m2 for 6–12 cycles) was administered to all patients at the time of management. Some patients received additional chemotherapies including etoposide, bevacizumab, irinotecan, and lomustine.
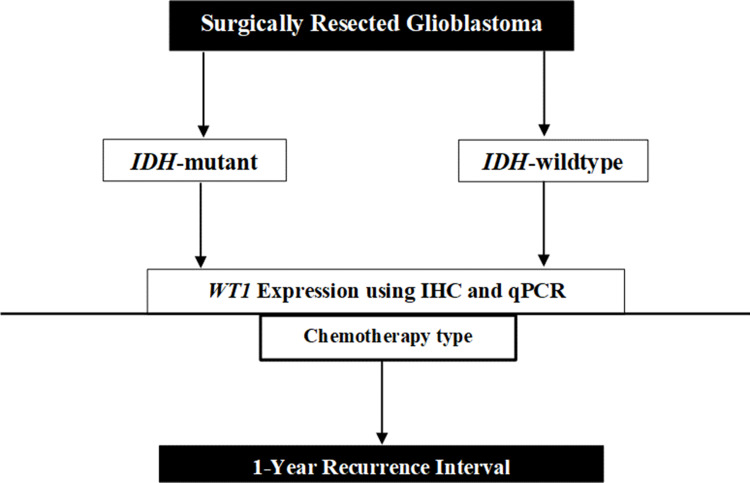
Figure 1.
Schematic of the approach used in this study. The samples have been categorized based on ISDH1 mutation and their WT1 expression. Recurrence interval used as determinant factor for patient’s outcome.
Immunohistochemistry Protocol Used for IDH1 Mutation Assessment
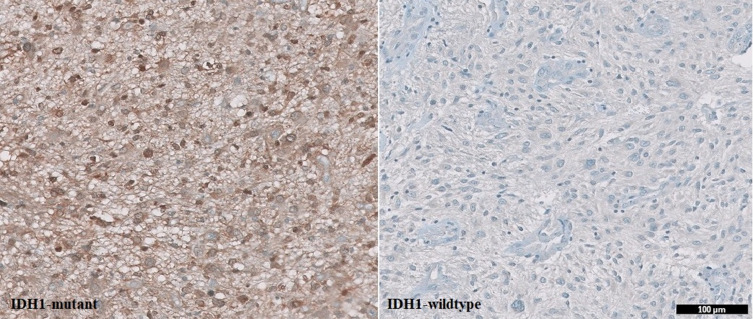
Anti-IDH1 antibody is intended for laboratory use to qualitatively identify IDH1 mutation in formalin-fixed paraffin-embedded tissue (FFPE) sections using an automated slide stainer. The IHC assay using anti-IDH1 R132H (Dianova, Clone H09) mouse monoclonal antibody was performed with an OptiView detection kit on a BenchMark XT (Ventana). The assay procedure consisted of deparaffinization with EZ Prep at 75°C, heat pretreatment with Cell Conditioner for 68 minutes, and incubation with 1:20–1:50 diluted antibody for 32 min at 37°C. Slides were counterstained with hematoxylin II and bluing reagent for 16 minutes. Sections in which >10% of tumor cells were positively stained were defined as mutant IDH1 (Figure 2).
Figure 2.
IDH1 mutation status in glioblastoma using immunohistochemistry (IHC). IDH1 mutation showed positive expression while IDH1-wildtype showed negative expression). Scale bar, 100 μm.
Assessment of WT1 Expression Through IHC and RT-qPCR
Assessment of WT1 Protein Expression Using IHC
Anti-Wilms tumor (WT1) antibody is intended for laboratory use to identify protein expression in FFPE sections on an automated slide stainer. The IHC assay using anti-WT1 (Clone 6F-H2, Ventana) mouse monoclonal antibody was performed using the DAB detection kit on a BenchMark XT (Ventana). The assay procedure consisted of deparaffinization with EZ Prep at 75°C, pretreatment with Cell Conditioner for 68 minutes, followed by incubation with 1:100–1:500 diluted antibody for 32 min at 37°C. Slides were removed from the slide stainer after counterstaining with hematoxylin II and were immersed into successive alcohol buffer for 3 min. Sections in which tumor cell cytoplasm was positively stained were defined as “WT1 expressed” by a certified neuropathologist.
Quantitative Analysis of WT1 Histochemical Expression on Glioblastomas
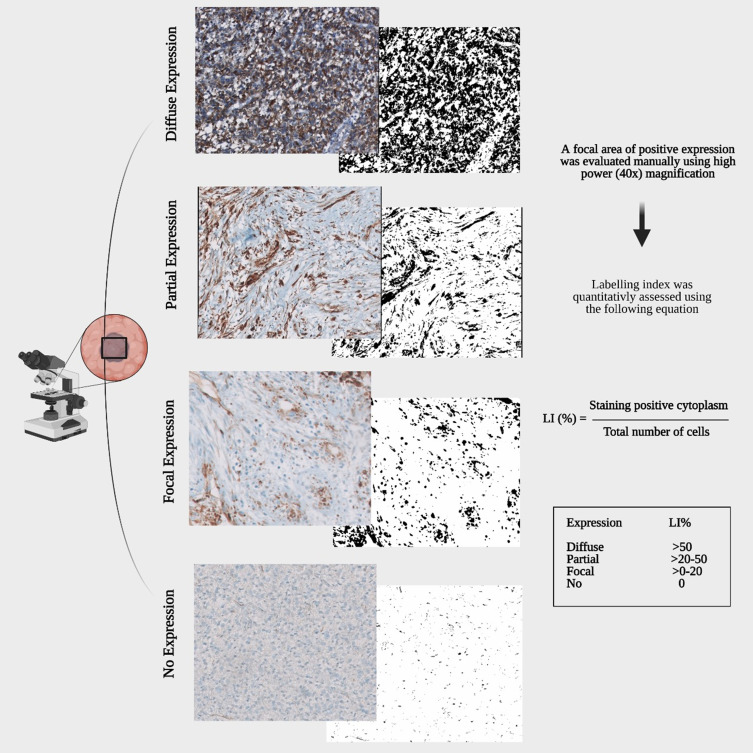
After immunostaining, a focal area of positive expression was evaluated under light microscopy using high-power (40×) magnification. Tumor cells were counted manually by a certified neuropathologist (MK). The labelling index was quantitatively assessed using the following equation: Labelling Index = [(Staining-positive cytoplasm)/(Staining-positive cytoplasm + Staining-negative cytoplasm)] (Table 1). The staining pattern was categorized as 1) overexpressed, 2) focal expressed, 3) partially expressed and 4) none-expressed (Figure 3).
Table 1.
The Labelling Index (%) Was Assessed Through the Following Scoring System
| Expression | Labelling Index (%) |
|---|---|
| No expression | 0 |
| Focal expression | >0–20 |
| Partial expression | >20–50 |
| Diffuse expression | >50 |
Note: For statistical analysis, the scores were divided by 100.
Figure 3.
An algorithm of analysis workflow describing the quantitative assessment of WT1 expression through IHC. The expression categories are based on labelling indices (%).
Assessment of WT1 Gene Expression Using Reverse Transcriptase-qPCR
H&E-stained sections from FFPE tissue blocks were examined by a neuropathologist (MK) to select regions from which RNA could be extracted. RNA was isolated by standard procedures from selected tissue fragments containing a high percentage of tumor cells. RNA extraction was performed using FFPE RNeasy Kit (no. 73504) according to the manufacturer’s instructions. CDNA was synthesized by ImProm-II™ Reverse Transcription System (CAT no. A3800). Quantitative-PCR was performed by QuantiFast SYBR® Green RT-PCR Kit (no. 204154) with two primer pairs, the targeted gene: WT1 and the reference gene: GAPDH (glyceraldehyde-3-phosphate dehydrogenase) (Table 2). The Annealing Temperature was 58C. Of the 44 enrolled glioblastoma cases, only 13 samples had adequate RNA quality for RT-PCR. This limitation should be taken into consideration during assessment of WT1 expression in pathological practice. The samples were divided into case (n = 13) and control (n = 2) groups. The control cases were low-grade glioma and non-glioma. IHC results showed no WT1 expression in control cases. Three replicates of threshold cycle (CT) values for five target genes and one reference gene were used for analysis. The mean CT and standard deviation for the reference (GAPDH) and target (WT1) genes were calculated from the RT-PCR data and analyzed by ∆∆CT and ∆CT methods. The average CT for the control and each tested gene was calculated from the data generated by RT-PCR using the Step One System and Data Assist software. The CT of the target gene was normalized to the CT of the reference gene, then the ∆CT of the test sample was normalized to the ∆CT of the control sample and the relative quantification (Rq) and differential expression (fold change, FC) were calculated using
Table 2.
Primers for WT1 Gene Expression Analysis
| Primer | Sequence |
|---|---|
| WT1-Forward | CACACGCACGGTGTCTTC |
| WT1-Reverse | AGATGCCGACCGTACAAG |
| GAPDH-Forward | CCCCCAATGTATCCGTTGTG |
| GAPDH-Reverse | TAGCCCAGGATGCCCTTTAGT |
Abbreviation: GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
(1) ΔCT for Ctrl or test = CT target gene – CT reference gene, (2) ΔΔCT = ΔCT test sample – ΔCT control sample, (3) Relative quantification (Rq) = 2 −∆∆CT = value*. The fold change (differential expression) was also calculated. ΔCT values for each sample were determined using the following formula: ΔCT = [mean CT reference gene – mean CT target gene; however, FC-WT1 >0 represents upregulation, whereas FC-WT1 <0 represents downregulation of gene (Table 3).
Table 3.
WT1 Gene Expression as Measured Using IHC and RT-qPCR
| WT1 Expression (IHC) | WT1 Expression (qPCR) | WT1 CT mean | GDPH CT mean | FCCT WT1 | Rq | |
|---|---|---|---|---|---|---|
| 1 | Overexpressed | Upregulated | 39.47 | 32.14 | 3.42 | 3.42 |
| 2 | Overexpressed | Upregulated | 36 | 29.19 | 4.9 | 4.9 |
| 3 | Partially expressed | Upregulated | 35.01 | 31.7 | 55.62 | 55.62 |
| 4 | Not expressed | Upregulated | 35.64 | 28.89 | 5.42 | 5.42 |
| 5 | Overexpressed | Upregulated | 36.53 | 27.65 | 1.17 | 1.17 |
| 6 | Overexpressed | Upregulated | 34.98 | 31.21 | 40.55 | 40.55 |
| 7 | Overexpressed | Downregulated | 37.5 | 19.9 | −361 | 0.0028 |
| 8 | Not expressed | Downregulated | 36.42 | 22.97 | −20.25 | 0.05 |
| 9 | Overexpressed | Downregulated | 37.5 | 23.8 | −24.49 | 0.04 |
| 10 | Overexpressed | Downregulated | 36.7 | 19.5 | −280 | 2.76 |
| 11 | Not expressed | Downregulated | 37.47 | 28.08 | −1.22 | 0.82 |
| 12 | Overexpressed | Downregulated | 38.95 | 21.3 | −374 | 0.0027 |
| 13 | Overexpressed | Downregulated | 39.98 | 23.25 | −197 | 0.0051 |
Abbreviations: CT, threshold cycle (CT); GAPDH, glyceraldehyde-3-phosphate dehydrogenase; FCCT, fold change cycle threshold; Rq, relative quantification.
Statistical Methods
Data are described as frequencies and percentages. The McNemar test was used to compare the sensitivity, specificity, and accuracy of IHC and RT-qPCR for WT1 gene expression detection. Kaplan–Meier curves were used to compare the distribution of recurrence-free interval (RFI) between mutant IDH1 and WT1 expression groups. Recurrence interval (RI) is defined as the period after total surgical resection to the first possible date of recurrence. All statistical analyses were performed using IBM SPSS1 ver. 24 statistical software programs (SPSS Inc., Chicago, IL).
Results
Forty-four patients with completely resected and treated glioblastoma were included in this study. The mean patient age was 54 years, with a male-to-female ratio 1.45. Parietal and frontal areas were the predominant tumor locations and tumors in these locations were observed in 33 cases (75%). IDH1wildtype was found in 26 cases (59.1%) and the remaining 18 cases (40.9%) were IDH1mutant. IHC revealed WT1 overexpression in 32 cases (72.7%), partial expression in 9 cases (20.5%), and no expression in 3 cases (6.8%). For the 13 cases in which WT1 expression was tested by qPCR, 6 and 7 cases had up- and downregulated WT1 expression, respectively (Tables 3 and 4). For post-surgical treatment, 41 patients (93.2%) received chemoradiotherapy and 3 patients did not receive any adjuvant therapies. Among patients who received chemotherapies, around 52% (n = 23) were treated with TMZ alone and 27% (n = 17) were treated with TMZ and additional chemotherapeutic agents. The mean recurrence interval was 579 days after the total surgical resection of the tumor. Approximately, 36% (n = 16) of the patients had tumor recurrence after 1-year of resection while 63.6% (n = 28) showed recurrence before 1-year of resection. Table 4 summarizes the descriptive distribution of the data.
Table 4.
Patients Data
| Overall (n=44) | |
| Age | |
| Mean (SD) | 54.8 (15.3) |
| Range | 11.0–82.0 |
| Gender | |
| Female | 18 (40.9%) |
| Male | 26 (59.1%) |
| Tumour Location | |
| Frontal | 16 (36.4%) |
| Occipital | 3 (6.8%) |
| Parietal | 17 (38.6%) |
| Temporal | 8 (18.2%) |
| IDH1 Status | |
| IDH-mutant | 18 (40.9%) |
| IDH-wildtype | 26 (59.1%) |
| WT1 Expression (IHC) | |
| Not expressed | 3 (6.8%) |
| Overexpressed | 32 (72.7%) |
| Partially expressed | 9 (20.5%) |
| Adjuvant Therapy | |
| Chemoradiotherapy | 41 (93.2%) |
| None | 3 (6.8%) |
| Chemotherapy Type | |
| Temozolomide | 23 (52.3%) |
| Other | 18 (41%) |
| Recurrence Interval | |
| Mean (SD) | 579.3 (348.8) |
| Range | 61.0–1344.0 |
| Recurrence Time | |
| <1 Year | 28 (63.6%) |
| >1 Year | 16 (36.4%) |
IHC and RT-qPCR Detection of WT1 Expression in Patients with Glioblastoma
There was no clear evidence in the literature whether IHC or qPCR showed better results when measuring WT1 expression. RT-qPCR can be processed using fragmented tissue, but the RNA content is often not enough to provide beneficial results. Therefore, we investigated the accuracy of both methods assuming that IHC, based on the previous published data, is more accurate.
Tumour samples (n=31) with low RNA concentration <20 nM have been excluded from qPCR test. IHC and qPCR were used in the 13 clustered samples that had RNA concentrations >20 nM. Two cases showed WT1 downregulation (no expression) in both IHC and qPCR. A single case, with low RNA concentration (22.7 gM) showed no WT1 expression by IHC but WT1 upregulation by qPCR. The five cases, that showed WT1 overexpression on IHC, showed downregulation by qPCR. The remaining five cases had parallel results. There was no statistically significant difference in WT1 expression between samples with different RNA concentrations (P-value >0.05). The McNemar test revealed that 83% sensitivity and 28.5% specificity were achieved using IHC rather than qPCR for assessing WT1 expression (Table 5). The lack of significant difference in WT1 expression between IHC and qPCR indicates that both methods are not reliable to be used in parallel.
Table 5.
McNemar Test Was Used to Detect Matching Compatibility Between Expression Results Obtained Using IHC and qPCR
| WT1 Expression Measured by qPCR | ||||
| WT1 Expression (IHC) | Downregulated | Upregulated | Total P-value | |
| Downregulated | 2 | 1 | 3 | 0.102x2 |
| Upregulated | 5 | 5 | 10 | 0. 221×2 c |
| Total | 7 | 6 | 13 | |
| Sensitivity | 83.3% | |||
| Specificity | 28.6% | |||
| Accuracy | 53.8% | |||
Relationship Between IDH1 Status, WT1 Expression, and Recurrence Interval
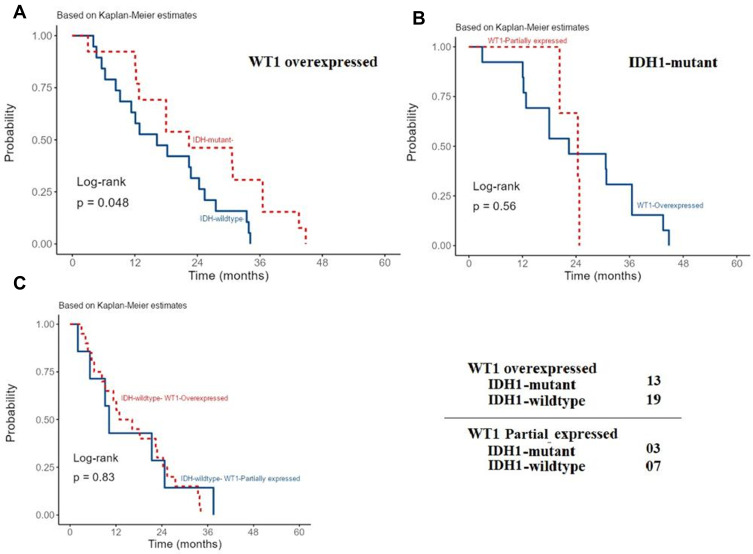
The recurrence interval among patients with WT1 overexpression significantly differs between cases with wild-type and mutant IDH1 (P-value=0.048). IDH1mutant cases showed late recurrence, after 1-year (Figure 4A). This significance was not observed among IDH1mutant cases with partially expressed or overexpressed WT1 (P-value = 0.56) (Figure 4B). These results indicate that in cases with mutant IDH1, WT1 upregulation lowers the chance of tumor recurrence. No statistically significant difference in recurrence interval was observed among IDH1wildtype cases with WT1 partial or overexpression (P-value = 0.83) (Figure 4C).
Figure 4.
Recurrence interval among patients with glioblastoma and different IDH1 status and WT1 expression. The recurrence interval among patients with WT1 overexpression significantly differs between wild-type and mutant IDH1 (P-value < 0.048) (A). This significance was not observed among cases with mutant IDH1 and partially expressed or overexpressed WT1 (P-value = 0.56) (B) as well as among cases with wild-type IDH1 and WT1 partial or overexpression (P-value = 0.83) (C).
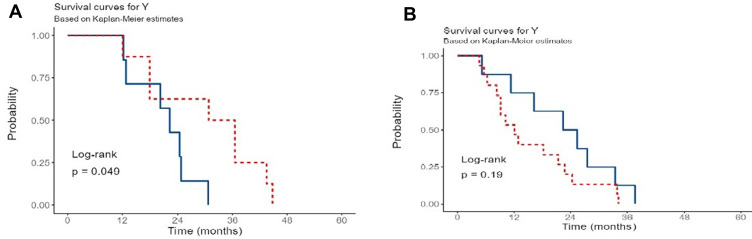
Moreover, IDH1mutant glioblastomas with WT1 overexpression, who received TMZ with additional chemotherapies, showed late recurrence intervals than those who received TMZ alone (P-value = 0.049). On the other hand, IDH1wildtype glioblastomas with WT1 overexpression who received TMZ or TMZ with additional chemotherapies showed no significant difference in RFI (P-value = 0.19) (Figure 5A and B)
Figure 5.
The recurrence interval among glioblastoma patients, with WT1 overexpression and different IDH1 status, who received different treatment modalities.
Notes: (A) The association between chemotherapies and IDH1 mutation (blue curve: IDH1 mutant with TMZ; red curve: IDH1 mutant with only TMZ and other chemotherapies). (B) The association between chemotherapies and wildtype IDH1 (blue curve: IDH1 wildtype with TMZ and other chemotherapies; red curve: IDH1 wildtype with only TMZ).
Discussion
Glioblastoma is the most aggressive primary malignant brain tumor in adults. While primary and secondary glioblastomas are pathologically indistinguishable, they vary at the molecular level. After surgical resection, the current standard treatment for patients with glioblastoma is radiotherapy and chemotherapy using either TMZ alone or TMZ with additional chemotherapeutic agents. The overall survival rate (OS) for patients with glioblastoma is around 14.6 months with a 5-year long-term survival. Nevertheless, glioblastoma remains a fatal disease, and treatment strategies are palliative.
WT1 encodes a zinc finger transcriptional factor, which has been implicated in various malignancies such as childhood renal neoplasm (WT), leukemias, breast and ovarian cancers.1–4 Recent studies have shown that WT1 plays a role in gliomagenesis.5 Its overexpression has been repetitively observed in high‑grade gliomas.4,6,7 However, the utility potential of WT1 expression as a biomarker has not been sufficiently substantiated. Testing WT1 gene or protein expression in patients with glioblastoma patients is important for treatment planning and prognostic determination. Recently, clinical trials of cancer immunotherapies targeting WT1 have shown promising results in glioblastomas, particularly in resistant cases, suggesting that WT1 is a potential target for immunotherapy, which increases the sensitivity of glioblastoma to chemotherapy.8
Although the immunohistochemical approach for assessing WT1 expression is a useful method, some studies have shown similar WT1 expression results using both IHC and molecular analyses.4 Our results showed that there was no significant difference in WT1 expression by using different tests (IHC or qPCR) with different RNA concentrations. However, low RNA volume and concentration may give false qPCR results. This does not occur when WT1 expression being tested by IHC. The proteins detected by IHC are more stable and well preserved after tissue processing. Indeed, it gives more accurate and sensitive results. On the other hand, the RNAs detected by Rt-PCR are not stable chemically and mostly degraded after tissue processing, especially FFPE tissue. Our analysis revealed that IHC has 83% sensitivity, 28.5% specificity, and 53% accuracy supporting the notion that IHC is a reliable test for WT1 expression. Moreover, our results showed that parallel testing of WT1 expression using IHC and qPCR was not reliable; thus, IHC provides more accurate results than qPCR.
The association between IDH1 mutation and WT1 expression has also not been thoroughly investigated. Manocha et al identified an inverse relationship between WT1 scores and IDH1 mutation.9 Rauscher et al found that some anaplastic astrocytomas and glioblastomas lack WT1 expression. They ascribed this lack of WT1 expression in high‑grade tumors to the younger age of patients and the presence of IDH1 mutations in the tumors.10 In TCGA dataset, 12 glioblastoma cases have been investigated. All the cases were IDH1wildtype while six cases had WT1 upregulated and six downregulated. The effect of WT1 expression on tumor recurrence interval was insignificant (p=value =0.149). In our analysis, we found that IDH1mutant glioblastomas with WT1 overexpression are associated with late tumor recurrence interval compared with cases with IDH1wildtype. The association between WT1 expression and the type of chemotherapy administered has also never been studied to date. Although WT1 immunotherapy is currently under trial, we found that TMZ taken with additional chemotherapies may improve the survival rate and decrease the chance of tumor recurrence in patients with glioblastomas. This means that TMZ with other chemotherapies may be used as an add-on to WT1 immunotherapy to prevent tumor regression, to increase the sensitivity to TMZ, and to decrease tumor recurrence.
Conclusion
Parallel testing for WT1 expression using IHC and qPCR is not reliable. However, IHC provides more accurate results. Moreover, IDH1-mutant glioblastomas with WT1 overexpression are associated with late RFI compared to IDH1 wild-type cases, particularly if temozolomide with additional chemotherapies are used.
Code Availability
N/A.
Funding Statement
The study is sponsored and funded by “Deanship of Scientific Research of King Abdulaziz University, Jeddah, Saudi Arabia” (Code No. G: 81-828-1441).
Data Sharing Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Ethical Approval
Ethical approval for this study was granted by the National Biomedical Ethics Committee at King Abdulaziz University (HA-02-J-008) (Reference No. 189-19). All procedures performed in this study involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards.
Consent to Participate
All contributors have given consent to participate in the study.
Author Contributions
MK, idea, IRB submission, writing, study design and data, histological analysis. NS, statistical analysis SB, data provider, writing, analysis AK, study design, statistical analysis, PCR analysis, writing, editing, YM, data entry, tissue collection, writing AB, data entry, tissue collection RS, data entry, tissue collection, writing BG, data analysis, editing, submission, consultation AL, data provider, IRB submission FM, tissue collection, IRB submission, IHC SH, data interpretation All authors contributed to data analysis, drafting or revising the article, have agreed on the journal to which the article will be submitted, gave final approval of the version to be published, and agree to be accountable for all aspects of the work.
Disclosure
All authors reported no conflicts of interest for this work.
References
- 1.Pritchard‑Jones K. The Wilms tumor gene, WT1, in normal and abnormal nephrogenesis. Pediatr Nephrol. 1999;13:620–625. doi: 10.1007/s004670050757 [DOI] [PubMed] [Google Scholar]
- 2.Call KM, Glaser T, Ito CY, et al. Isolation and characterization of a zinc finger polypeptide gene at the human chromosome 11 Wilms’ tumor locus. Cell. 1990;60:509–520. doi: 10.1016/0092-8674(90)90601-A [DOI] [PubMed] [Google Scholar]
- 3.Nakatsuka S, Oji Y, Horiuchi T, et al. Immunohistochemical detection of WT1 protein in a variety of cancer cells. Mod Pathol. 2006;19:804–814. doi: 10.1038/modpathol.3800588 [DOI] [PubMed] [Google Scholar]
- 4.Schittenhelm J, Beschorner R, Simon P, et al. Diagnostic value of WT1 in neuroepithelial tumor. Neuropathol Appl Neurobiol. 2009;35:69–81. doi: 10.1111/j.1365-2990.2008.00957.x [DOI] [PubMed] [Google Scholar]
- 5.Clark AJ, Ware JL, Chen MY, et al. Effect of WT1 gene silencing on the tumorigenicity of human glioblastoma multiforme cells. J Neurosurg. 2010;112:18–25. doi: 10.3171/2008.11.JNS08368 [DOI] [PubMed] [Google Scholar]
- 6.Kijima N, Hosen N, Kagawa N, et al. Wilms’ tumor 1 is involved in tumorigenicity of glioblastoma by regulating cell proliferation and apoptosis. Anticancer Res. 2014;34:61–67. [PubMed] [Google Scholar]
- 7.Somasundaram A, Ardanowski N, Opalak CF, et al. Wilms tumor 1 gene, CD97, and the emerging biogenetic profile of glioblastoma. Neurosurg Focus. 2014;37:1–4. doi: 10.3171/2014.9.FOCUS14506 [DOI] [PubMed] [Google Scholar]
- 8.Izumoto S, Tsuboi A, Oka Y, et al. Phase II clinical trial of Wilms tumor 1 peptide vaccination for patients with recurrent glioblastoma multiforme. J Neurosurg. 2008;108:963–971. doi: 10.3171/JNS/2008/108/5/0963 [DOI] [PubMed] [Google Scholar]
- 9.Manocha A, Jain S. WT1 in astrocytomas: comprehensive evaluation of immunohistochemical expression and its potential utility in different histological grades. Indian J Cancer. 2019;56:197–201. doi: 10.4103/ijc.IJC_51_18 [DOI] [PubMed] [Google Scholar]
- 10.Rauscher J, Beschorner R, Gierke M, et al. WT1 expression increases with malignancy and indicates unfavourable outcome in astrocytoma. J Clin Pathol. 2014;67:556–561. doi: 10.1136/jclinpath-2013-202114 [DOI] [PubMed] [Google Scholar]