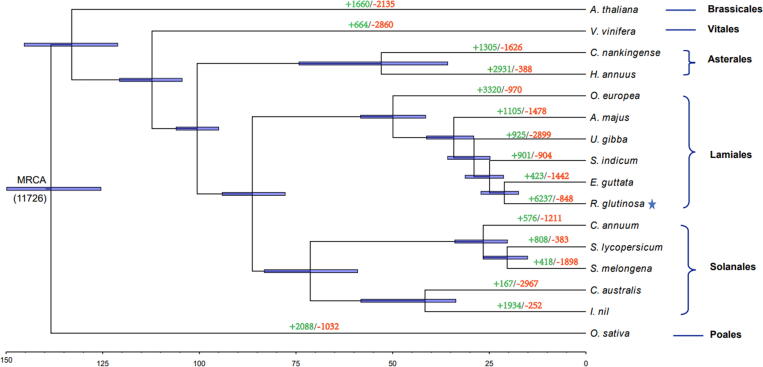
Fig. 3.
Phylogenetic analysis and divergence time estimations among 16 plant species including R. glutinosa. The tree was constructed based on all single-copy orthologous genes using Oryza sativa as outgroup. A total of 500 bootstrap replicates was performed and bootstrap values lower than 100 are not presented. The star marks the species used for genome sequencing in this study. Divergence times estimated in million years ago are indicated by the blue lines over the nodes. The span of the blue lines shows 95% confidence interval of the divergence time. The divergence time was estimated for each node in million years (MY). The number of gene-family contraction and expansion events is indicated by green and red numbers, respectively. The number at the root (11,726) denotes the total number of gene families predicted in the most recent common ancestor (MRCA). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)