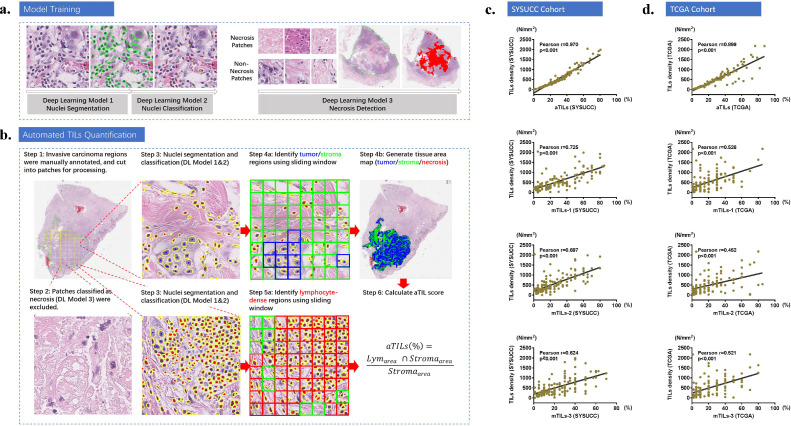
Fig. 1.
Overview of the deep learning model used for CTA and the correlation between TIL density and TIL scores. (a) Three deep learning models were trained for nuclei segmentation, nuclei classification, and necrosis detection. (b) A flowchart of the automated stromal TIL quantification process. Step 1: The invasive tumor area was manually outlined by the pathologist (green contour). Step 2: The regions inside the outlined area were cut into 224×224 pixels patches and classified into necrotic and non-necrotic; the necrotic patches were excluded for the following analysis. Step 3: The nuclei in each non-necrosis patch were segmented (yellow contours) and classified into tumor (blue dots), lymphocyte (red dots) and others (no dot). Step 4: A sliding window of 128×128 pixels corresponding to a size of 32×32 µm was used to visualize all of the non-necrotic regions; if the number of tumor cell nuclei in the window was greater than 2, then this region was considered a tumor region (blue blocks). The remaining regions were recognized as the stromal region (green blocks). The identified tumor, stroma and necrosis regions in the entire WSI were also shown. Step 5: A smaller 32×32 pixel (8 × 8 µm) sliding window was used to view all of the stromal regions; if the number of lymphocytes in the window was greater than 2, then the region was considered lymphocyte-dense (red blocks). Step 6: The final aTIL score was calculated as the overlapping area between lymphocyte-dense regions and stromal regions divided by the area of the stromal regions. The sliding window without overlap was shown for illustration purpose, as 50% overlapping was actually adopted. (c, d) The stromal TIL density within the invasive tumor area positively correlated with the mTIL and aTIL scores in both cohorts.