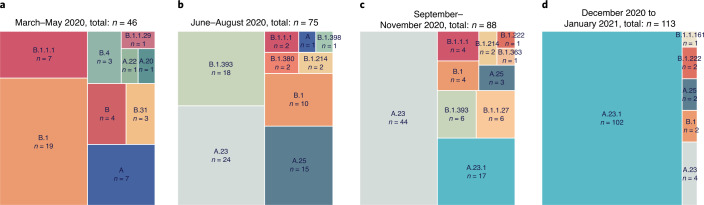
Fig. 1. SARS-CoV-2 lineage diversity in Uganda.
All high-coverage complete sequences from Uganda (n = 322) were lineage-typed using the pangolin resource (https://github.com/cov-lineages/pangolin). Lineage counts were stratified into four periods: March–May 2020 (a); June–August 2020 (b); September–November 2020 (c); and December 2020 to January 2021 (d). The percentage of each lineage within each set was plotted as a treemap using squarified treemap41 implemented in squarify (https://github.com/laserson/squarify) with the size of each sector proportional to the number of genomes; genome numbers are listed with ‘n = ’.