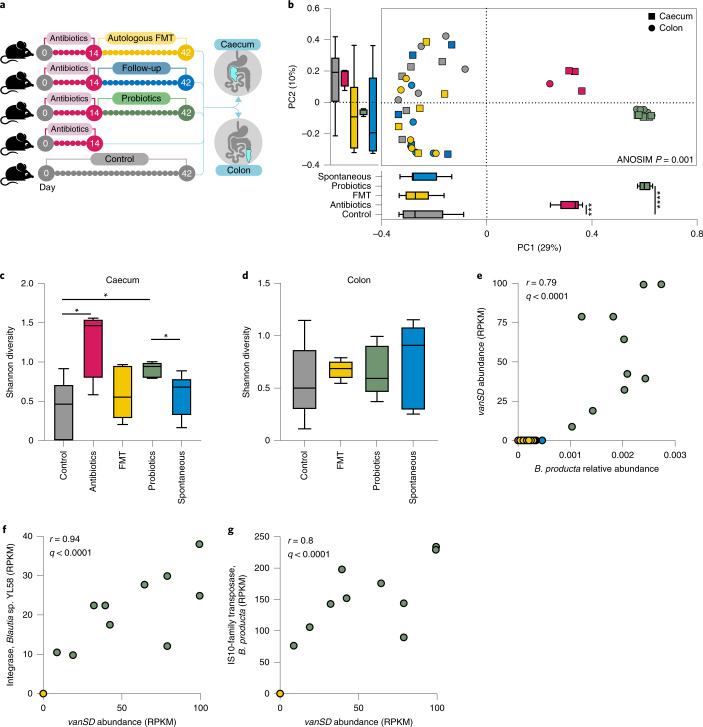
Fig. 5. Probiotics expand the GI tract resistome in antibiotic-treated mice.
a, Experimental design. Wild-type adult (10-week-old) male C57BL/6J mice were treated with ciprofloxacin and metronidazole in their drinking water for 2 weeks followed by either daily supplementation by oral gavage with a probiotic supplement (Bio-25; green), autologous FMT performed after the last day of antibiotics (yellow) or spontaneous recovery (blue). The three groups were killed after 28 d of recovery and a fourth group was killed immediately after antibiotics (magenta). A fifth control group was untreated throughout the 42-d experimental period (grey). From the original experiment, which included 10 mice per group, we randomly selected 5 mice (spanning both cages per group) and performed shotgun metagenomic sequencing and resistome profiling of caecal and distal colon luminal content using ShortBRED and CARD after subsampling to 1.5 M of reads. Results are based on ARG families. b, Bray–Curtis dissimilarities. PC1 antibiotics versus control P = 0.0317, probiotics versus control P = 0.0317, probiotics versus spontaneous P = 0.0317. c,d, Shannon alpha diversity in the caecum lumen (c) or distal colon lumen (d). The antibiotics group was not included in the distal colon panel because four samples were under the subsampling threshold. c, Probiotics versus spontaneous recovery P = 0.0317; probiotics versus control P = 0.0317; control versus antibiotics P = 0.0317. e, Abundance of the vanSD gene cluster in the different groups and its Spearman correlation with B. producta abundance. f,g, MGEs significantly (P < 0.0001) correlated (Spearman) with vanSD abundance. f, Integrase, Blautia sp. YL58. g, IS-10 family transposase, B. producta. *P < 0.05; ***P < 0.001; ****P < 0.0001, two-sided Mann–Whitney U-test. The horizontal lines represent the median and the whiskers represent the 10–90 percentiles. RPKM, reads per kilobase of reference sequence per million sample reads.