Table 1.
Leishmania tarentolae aminoacyl-tRNA synthetases: gene annotation and proposed mechanisms for the formation of the cytosolic and the mitochondrial isoforms
| TriTrypDB ID | Name | MTSa probability MITOPROT | Location of predicted MTS | 5′-UTR size (bp) C | 5′-UTR size (bp) M | Number of reads | Comments |
|---|---|---|---|---|---|---|---|
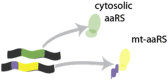 Cytosolic and mitochondrial aaRSs encoded by two distinct genes Cytosolic and mitochondrial aaRSs encoded by two distinct genes | |||||||
| LtaP30.0530 | AspRS C | / | / | 163 | / | 91 | |
| LtaP21.1070 | AspRS M | 0.85 | N-term | / | 176 | 20 | Start AUG misannotated in TriTrypDB |
| LtaP15.0260 | LysRS C | / | / | 89 | / | 65 | |
| LtaP30.0190 | LysRS M | 0.99 | N-term | / | 168 | 4 | |
| LtaP29.0060 | TrpRS C | / | / | 100 | / | 78 | Start AUG misannotated in TriTrypDB |
| LtaP23.0360 | TrpRS M | 0.95 | N-term | / | 136 | 393 | |
| LtaP29.2400 | ProRS C | / | / | 306 | / | 4 | Incomplete sequence in TriTrypDB |
| ? (LtaP18.1200) | ProRS M | 0.96 | N-term | / | 80 | 8 | Incomplete sequence in TriTrypDB |
 Cytosolic and mitochondrial aaRSs encoded a single gene Cytosolic and mitochondrial aaRSs encoded a single gene | |||||||
Verified alternative trans-splicingb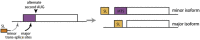
| |||||||
| LtaP34.2190 | AsnRSc | 0.75 | N-term | 82 | 148 | C61/M1 | Major splice site leads to cytosolic isoform, and the alternate minor splice site leads to the mitochondrial isoform |
| LtaP15.1380 | GlnRSc | 0.21 | N-term | 139 | 44 | C54/M1 | |
| LtaP36.5770 | IleRSc | 0.58 | N-term | 229 | nd | C2/M0 | |
| LtaP30.0700 | HisRS | 0.64 | Internal | 68 | 27 | C61/M1 | |
Presence of a predicted internal MTSd (hypothetical mechanism)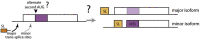
| |||||||
| LtaP27.1400 | ArgRSc | 0.64 | Internal | 107 | nd | 27 | Major splice site likely leads to the cytosolic isoform |
| LtaP11.0110 | SerRSc | 0.37 | Internal | 139 | nd | 29 | |
| LtaP13.1000 | LeuRSc | 0.61 | Internal | 90 | nd | 18 | |
| LtaP12.0270 | CysRSc | 0.74 | Internal | 259 | nd | 29 | |
| LtaP22.1510 | AlaRS | 0.99 | Internal | 249 | nd | 28 | |
| LtaP30.3140 | ValRS | 0.83 | Internal | 292 | nd | 16 | |
| LtaP35.1480 | ThrRS | 0.62 | Internal | 526 | nd | 7 | |
| LtaP36.3950 | GlyRS | 0.88 | Internal | 245 | nd | 29 | |
| LtaP32.0940 | PheRSα | 0.99 | Internal | 167 | nd | 29 | |
| LtaP19.0950 | PheRSβ | 0.91 | Internal | 252 | nd | 3 | |
| Unpredictable mechanism | |||||||
| LtaP30.3280 | GluRS | 0.62 | N-term | ? | 21 | 92 | Major splice site likely leads to the mitochondrial isoform |
| LtaP21.0830 | MetRS | 0.30 | N-term | ? | 227 | 20 | No alternative AUG predicted |
| LtaP14.1430 | TyrRS | Not predicted | Not predicted | 197 | ? | 83 | |
Abbreviations: C, cytosolic; M mitochondrial; nd, not detected.
Sequences for Leishmania tarentolae aminoacyl-tRNA synthetases (aaRSs) obtained from our nanopore RNA-Seq experiments are compared with those proposed in the TriTrypDB (https://tritrypdb.org/tritrypdb/ (26)), for which the ID is listed in the left column. TriTrypDB IDs in gray correspond to sequences that are either misannotated or incomplete in the database. Corrected sequences are provided in Supporting information.
MTS probability is predicted on the translated protein sequences by the MITOPROT software (39).
Both alternatively spliced mRNAs were detected in our RNA-seq data with the exception of the IleRS for which only the shorter cytosolic form was sequenced.
The same mechanism as described by Rettig et al. (20) or Nilsson et al. (18) in Trypanosoma brucei.
The possibility of an internal MTS is also predicted using the MITOPROT software. No experiments were conducted to validate these predictions.
