Fig. 3. VAX1 regulates transcription from the KISS1 promoter in KTaR-3 cells.
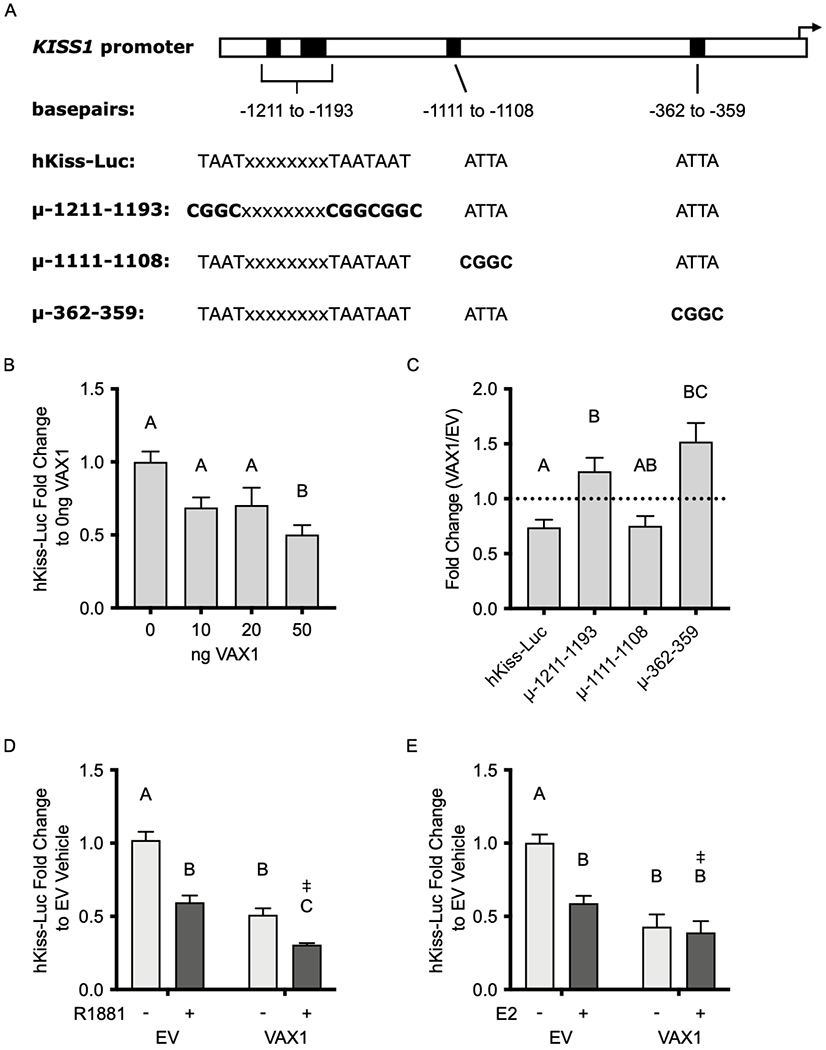
(A) Schematic of ATTA binding sites (black rectangles) on the KISS1 promoter and the sequence of these sites on the hKiss-Luc reporter or hKiss-Luc constructs containing cis-mutations at the indicated positions upstream of the transcriptional start site (black arrow). Bolded sequences indicate base pairs that were changed via site-directed mutagenesis. (B) hKiss-Luc was co-transfected with increasing concentrations of VAX1 expression vector into KTaR-1 cells. Data are represented as fold change as compared to 0 ng VAX1. (C) hKiss-Luc, μ-1211-1193, μ-1111-1108, and μ-362-359 were co-transfected into KTaR-1 cells with 50 ng VAX1 or EV. Data is represented as fold change of VAX1/EV. Dotted line indicates EV/EV. (D) KTaR-1 cells were transfected with hKiss-Luc or pGL2, 50 ng VAX1 or empty vector (EV), and 100 ng AR plasmids, and then treated with 10 μM R1881 (+) or vehicle (−). Data are represented as fold change as compared to EV vehicle. (E) KTaR-1 cells were transfected with hKiss-Luc or pGL2, 50 ng VAX1 or EV, and 50 ng ERα plasmids, and then treated with 1 pM E2 (+) or vehicle (−). Data are represented as fold change to EV vehicle. For all experiments, values represent means ± SEM. Data were analyzed using (B, C) One- or (D, E) Two-way ANOVA. Different letters denote significance, p<0.05. ‡ denotes a significant two-way factor interaction, p<0.05. N=3-4.
