Figure 1. Rps6 haploinsufficiency in the developing limb bud mesenchyme leads to selective patterning defects marked by p53 activation and reduced global protein synthesis.
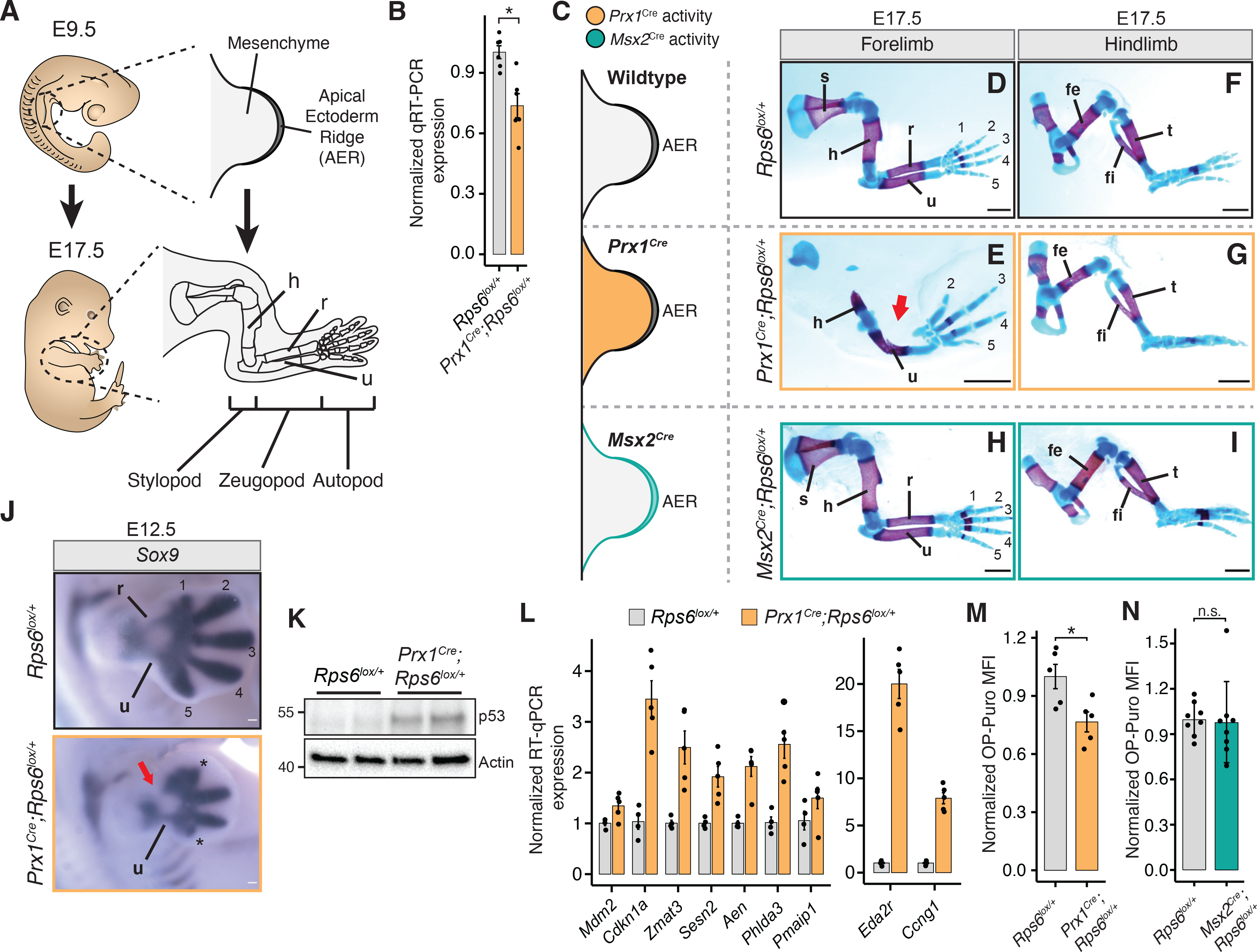
(A) Overview of forelimb development. AER, apical ectodermal ridge; h, humerus; r, radius; u, ulna.
(B) RT-qPCR of Rps6 mRNA from whole E10.5 forelimbs.
(C) Cre recombinase activity distribution in limb bud mesenchyme (Prx1Cre; orange), or in the overlying AER (Msx2Cre; green).
(D-I), E17.5 forelimbs and hindlimbs from WT (top row), Prx1Cre;Rps6lox/+ (middle row), and Msx2Cre;Rps6lox/+ (bottom row). Bone, red; Cartilage, blue. Numbers indicate digits. Arrow indicates absence of radius. Scale bars, 1 mm.
(J) Sox9 in situ hybridization of E12.5 forelimbs. Numbers indicate mesenchymal digit condensations. *Impaired digit development. Arrow indicates absence of radius. Scale bars, 0.1 mm.
(K) Representative p53 Western blot in whole E10.5 forelimbs. n = 2 embryos each.
(L) RT-qPCR of p53 target genes in whole E10.5 forelimbs. n = 4 embryos (Rps6lox/+), n = 5 embryos (Prx1Cre;Rps6lox/+).
(M) OPP MFI of cells dissociated from whole E10.5 Prx1Cre;Rps6lox/+ forelimbs normalized to WT. n = 5 embryos; MFI = median fluorescence intensity.
(N) OPP MFI of E11.5 Msx2Cre;Rps6lox/+ ectodermal cells normalized to WT. n = 8 embryos.
