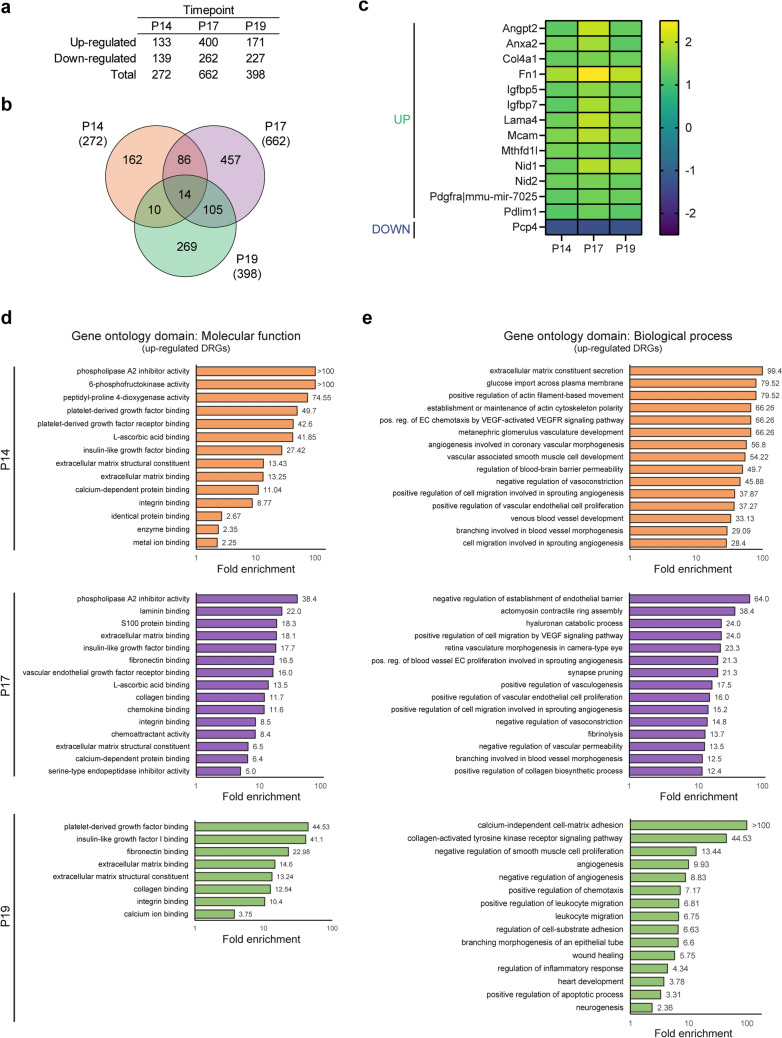
Figure 3.
Differential gene expression and GO term enrichment analyses on retinas of OIR mice relative to retinas of normoxic age-matched controls at each of the assessed timepoints. (a) Differential gene expression analysis on OIR retinas compared to normoxic age-matched controls across timepoints. Important deviations in gene expression profiles are detected, the most prominent of which occurs at P17. (b) Venn diagram of differentially expressed genes in OIR retinas relative to normoxic age-matched control samples. Only 14 transcripts were identified as differentially regulated at all three timepoints. (c) Heatmap depicting the fold-change expression of the 14 differentially regulated genes identified in (b), of which 13 were up-regulated and 1 was down-regulated. Noteworthy, the expression levels of up-regulated genes remained high throughout time, while the expression of the down-regulated transcript remained low at all timepoints examined. (d) GO term enrichment analysis in the Molecular function domain was conducted using all differentially up-regulated genes identified in OIR retinas relative to normoxic retinas at each time point (P14, top; P17, middle; P19, bottom). A subset of all enriched GO terms and their fold enrichment are depicted. (e) GO term enrichment analysis in the Biological process domain was conducted using all differentially up-regulated genes identified in OIR retinas relative to normoxic retinas at each time point (P14, top; P17, middle; P19, bottom). A subset of all enriched GO terms and their fold enrichment are depicted.