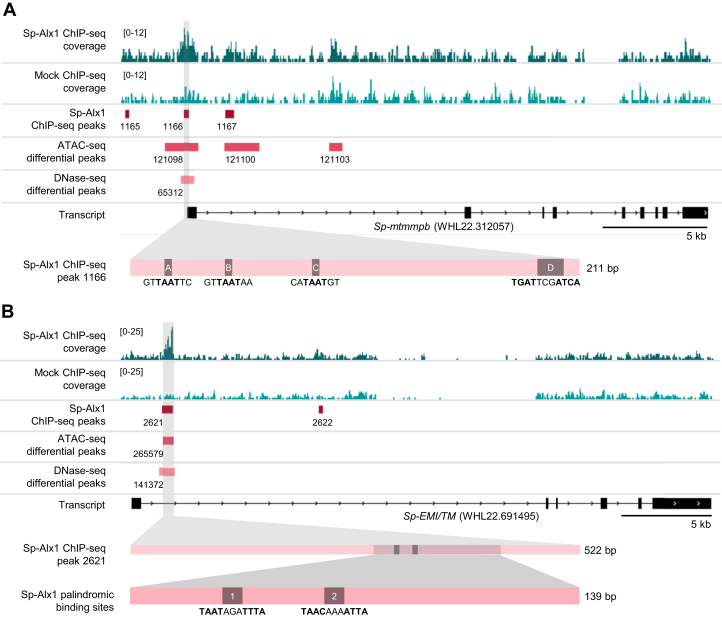
Figure 1.
Sp-Alx1 ChIP-seq peaks in the Sp-mtmmpb CRM used to identify Alx1-binding sites.A, genome tracks showing the location of the 210-bp Sp-Alx1 ChIP-seq peak (1166) in the 5′ untranslated region of Sp-mtmmpb, overlapping ATAC-seq and DNase-seq differential peaks (i.e., regions of chromatin selectively accessible in skeletogenic PMCs, cells that specifically express Sp-mtmmpb). Four putative Alx1-binding sites are present within ChIP-seq peak 1166. B, previously identified Sp-Alx1 ChIP-seq peak within an intron of Sp-EMI/TM. This region drives PMC-specific reporter expression in transgenic embryos, and expression depends on the first palindromic site (7). The DNA sequence of the first site was used in the EMSA assays. CRM, cis-regulatory module; PMCs, primary mesenchyme cells.