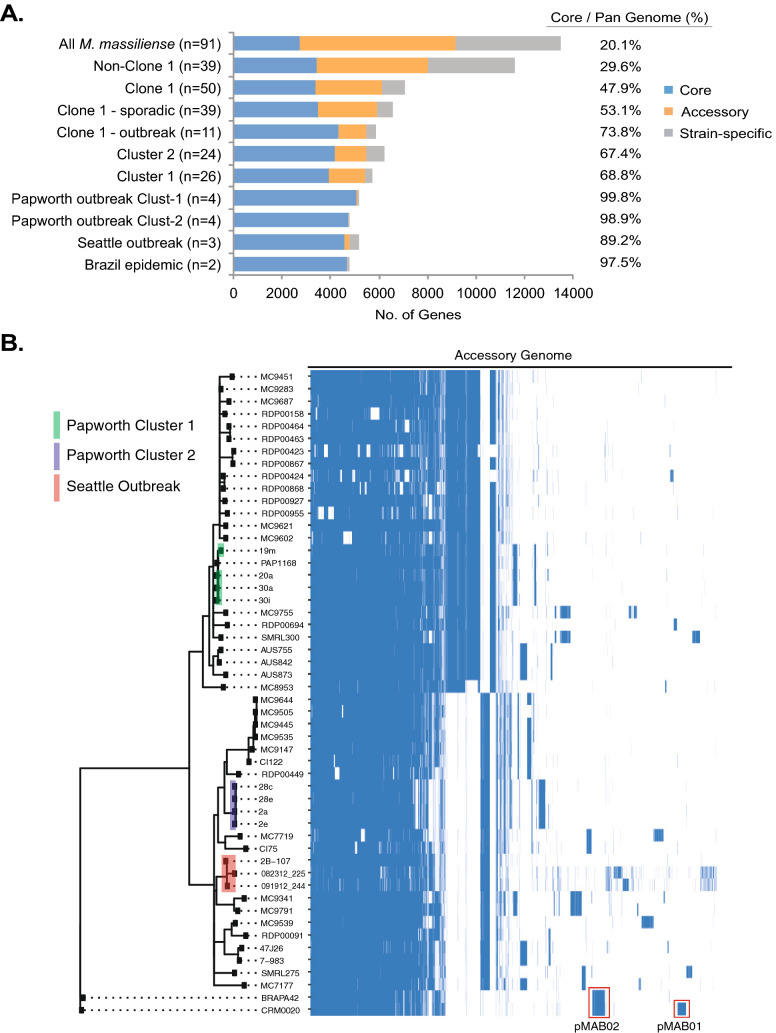
Figure 3.
Pan genome of M. massiliense. Pan genome analyses were performed for M. massiliense isolates that passed genome assembly quality filters (see “Methods”; n = 91). (A) Pan genome results for all isolates in the dataset and various isolate subgroups are shown as numbers of core genes, accessory genes and strain-specific genes. Percent of core genes for each group is shown (core genes/ total genes in pan genome). (B) Visualization of the accessory genome for Clone 1 M. massiliense isolates. Accessory genes for Clone 1 isolates (n = 50) and Brazilian epidemic isolates (n = 2) are shown as a presence or absence heatmap (blue means presence of a gene). Samples are ordered based on the core genome phylogenetic tree (left side). Known outbreak isolate clades are color coded for reference, and plasmids are indicated with red boxes.