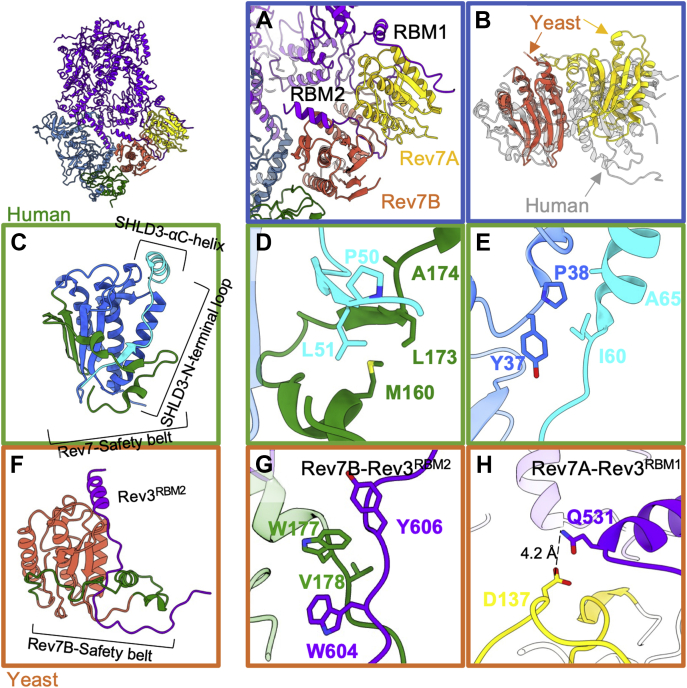
Figure 3.
Comparison of yeast and human Rev7 dimerization states and binding modes.A, Rev7 dimer in the yeast apo Polζ complex. Rev7A (yellow) and Rev7B (orange) bind to Rev7-binding motif 1 (RBM1) and 2 (RBM2) of Rev3 (purple), respectively. The RBM2 motif is sandwiched between the two Rev7 protomers, limiting its structural mobility. The RBM1 motif is more accessible than RBM2 suggesting less spatial restriction. B, overlay of yeast and human Rev7 dimers with yeast Rev7B (orange) and one of the human Rev7 protomers (gray) oriented in the same manner. This comparison highlights the radically different orientations of the two protomers in yeast (orange and yellow) and human (gray) Rev7. C, crystal structure of human Rev7(R124A) in complex with SHLD3 (41–74 aa). Rev7 core, safety belt region, and SHLD3 are shown in blue, green and cyan, respectively. D, safety belt region of human Rev7 (M160, L173 and A174) interacts with the N-terminal loop of SHLD3 (P50 and L51). E, interaction between human Rev7 (Y37 and P38) and SHLD3 (I60 and A65). F, structure of yeast Rev7B-Rev3RBM2 within the apo Polζ complex. Rev7B, safety belt region, and Rev3RBM2 are shown in orange, green and purple, respectively. G, interaction between the safety belt of yeast Rev7B (W177 and V178) and Rev3RBM2 motif (W604 and Y606). H, interaction between yeast Rev7A (D137) and Rev3RBM1 (Q531). Rev7A is colored yellow.