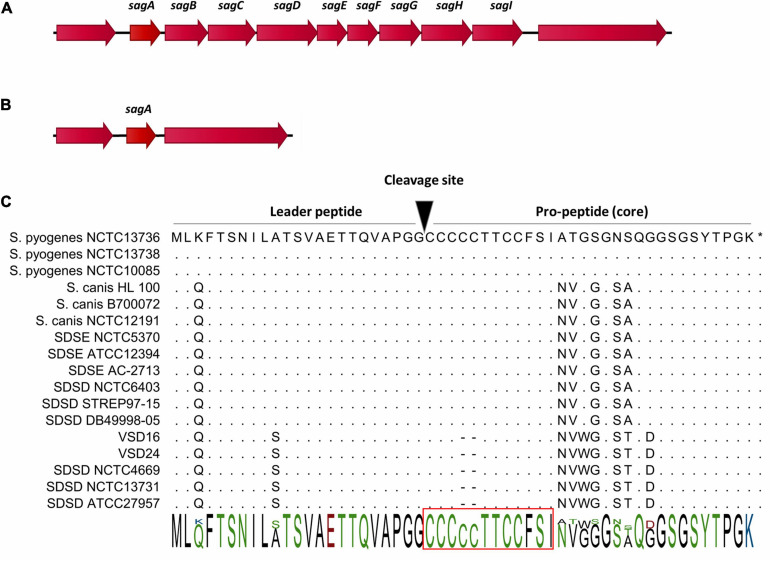
FIGURE 3.
Gene cluster organization of streptolysin S (SLS) operon and precursor peptide sequences of SLS cytolysins (SagA). (A) Organization of the SLS operon in Streptococcus dysgalactiae subsp. equisimilis (SDSE), human and fish S. dysgalactiae subsp. dysgalactiae (SDSD), Streptococcus pyogenes, and Streptococcus canis. The operon encoding SLS includes the prepropeptide structural gene (sagA), followed by eight genes responsible for the conversion of SagA into SLS (sagBCD), transport across the membrane (sagFGHI), and accountable for leader cleavage (sagE) (Datta et al., 2005). (B) Organization of the genomic region flanking sagA in bovine SDSD isolates. (C) Alignment of SagA peptide, the precursor of SLS. SLS core region possesses a highly conserved N-terminus, while the C-terminus is more variable. The putative leader peptide cleavage site is shown. Red rectangle, minimal core region required for hemolytic activity of SLS in S. pyogenes. STAB-Vida performed sequencing of sagA. Deduced amino acid sequences from this bovine allele were compared with sequences from the National Center for Biotechnology Information (NCBI) database and were analyzed with the CLC-bio Main Workbench sequence alignment tool (QIAGEN, Netherlands).