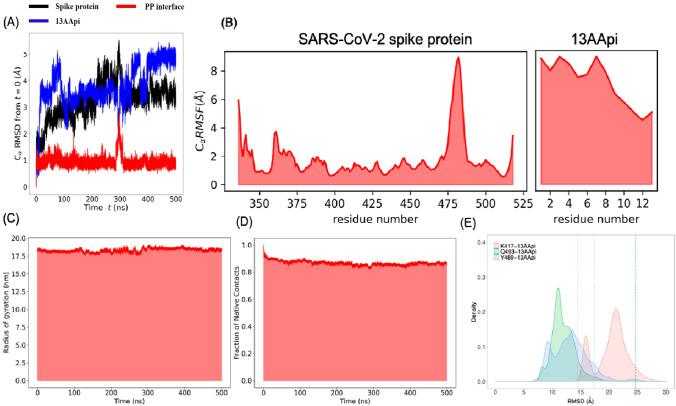
Fig. 3.
MD simulation to study the dynamics of the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2) spike protein and 13-amino acid peptide inhibitor (13AApi) complex. A Root mean squared deviation (RMSD) of the Cα atoms of the SARS-CoV-2 spike protein (black), 13AApi (blue), and protein-peptide interface (red) plotted against total simulation time. B Root mean squared fluctuation of SARS-CoV-2 spike protein (black) and 13AApi. C The radius of gyration of the whole protein-peptide complex. D Fraction of native contacts of the whole protein-peptide complex. E Distance between Cα atom of K417, Y489, and Q493 with the center of mass of 13AApi at t = 0 ns is shown as dashed lines