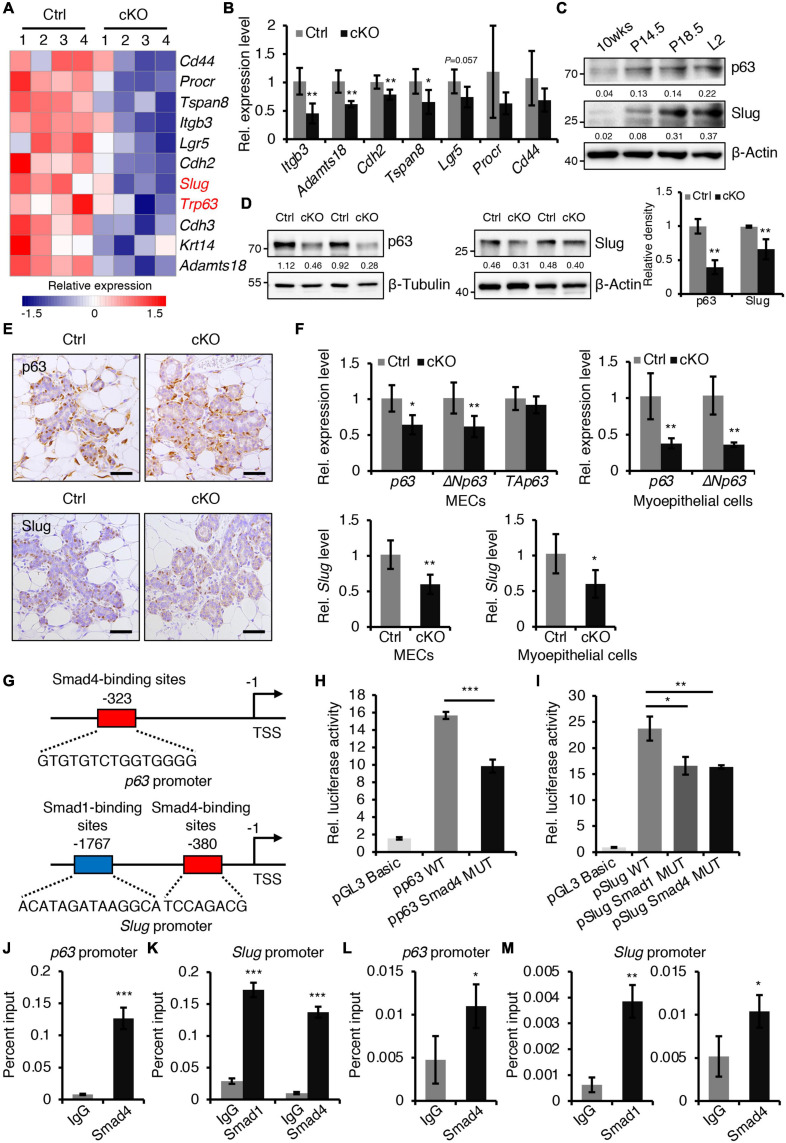
FIGURE 5.
BMPR1a regulated the expression of p63 and Slug through the pSmad1/5-Smad4 complex. (A) Heatmap for myoepithelial layer-related gene expression in control and cKO mammary glands at pregnancy day 14.5. n = 4 mice per group. (B) qRT-PCR analysis of Itgb3, Adamts18, Cdh2, Tspan8, Lgr5, Procr, and Cd44 in FACS-sorted myoepithelial cells from control and cKO mice at pregnancy day 14.5. n = 4–5 biological replicates. (C) Western blotting for p63 and Slug in wild-type mouse mammary glands at the indicated time points (P14.5, pregnancy day 14.5; P18.5, pregnancy day 18.5; L2, lactation day 2). β-Actin was used as a loading control. (D) Western blotting for p63 and Slug in mammary epithelial cells isolated from control and cKO mice at pregnancy day 14.5. β-Tubulin and β-Actin were used as loading controls. Statistical analysis the expression of p63/β-Tubulin (n = 3 mice) and Slug/β-Actin (n = 4 mice). (E) Immunohistochemistry staining for p63 and Slug in control and cKO mammary glands at pregnancy day 14.5. n = 4 mice per group. Scale bar, 50 μm. (F) qRT-PCR analysis of p63,ΔNp63, TAp63, and Slug in isolated mammary epithelial cells (MECs) and FACS-sorted myoepithelial cells from control and cKO mice at pregnancy day 14.5. n = 4-6 biological replicates. (G) Schematic diagram showing potential Smad4-binding sites (–323 bp) in the p63 promoter and potential Smad1-binding sites (–1767 bp) and Smad4-binding sites (–380 bp) in the Slug promoter. (H) Luciferase activity in lysates of HC11 cells transfected with pGL3-basic empty vector, wild-type p63 promoter (control), or mutant promoter (with mutation of Smad4-binding sites) luciferase reporter plasmids. n = 3 biological replicates. (I) Luciferase activity in lysates of HC11 cells transfected with pGL3-basic empty vector, wild-type Slug promoter (control), or mutant promoter (with mutation of Smad1- or Smad4-binding sites) luciferase reporter plasmids. n = 3 biological replicates. (J) ChIP analysis for the binding sites of Smad4 in the p63 promoter in HC11 mammary epithelial cells treated with BMP4 for 1 h using antibodies against Smad4. IgG was used as a negative control. The enrichment of Smad4 binding to p63 promoter was quantified using qPCR. n = 3. (K) ChIP analysis for the binding sites of Smad1 or Smad4 in the Slug promoter in HC11 mammary epithelial cells treated with BMP4 for 1 h using antibodies against Smad1 or Smad4. IgG was used as a negative control. The enrichment of Smad1 or Smad4 binding to Slug promoter was quantified using qPCR. n = 3. (L) ChIP analysis for the binding sites of Smad4 in the p63 promoter in primary mammary epithelial cells isolated from mouse mammary glands at pregnancy day 14.5 using antibodies against Smad4. IgG was used as a negative control. The enrichment of Smad4 binding to p63 promoter was quantified using qPCR. n = 3. (M) ChIP analysis for the binding sites of Smad1 or Smad4 in the Slug promoter in primary mammary epithelial cells isolated from mouse mammary glands at pregnancy day 14.5 using antibodies against Smad1 or Smad4. IgG was used as a negative control. The enrichment of Smad1 or Smad4 binding to Slug promoter was quantified using qPCR. n = 3. Data were presented as means ± SD. ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001.