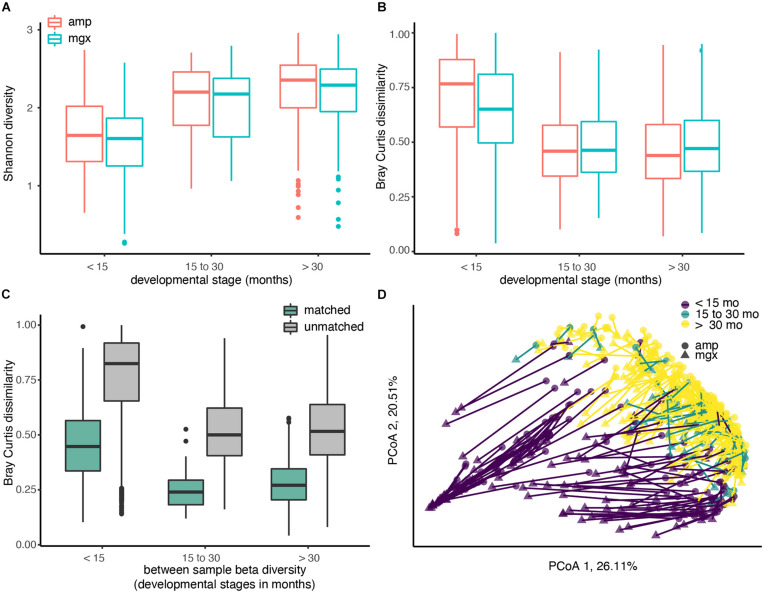
FIGURE 1.
Diversity of the child gut microbiome differs by age, regardless of profiling method. Microbiome communities from 338 children were sequenced using 16S rRNA (abbreviated “amp”) and shotgun metagenomic (abbreviated “mgx”) profiling. (A) Alpha diversity was calculated using the Shannon diversity index for each child. Boxplots are grouped by age and colored by profiling method. (B) Beta-diversity was quantified using pair-wise Bray-Curtis dissimilarities between all children within the same profiling method and developmental stage. (C) Bray-Curtis dissimilarities between 16S and metagenomic profiles for matched samples (from same fecal sample), 16S and metagenomic profiles among unmatched samples (from different fecal samples). (D) Beta-diversity was visualized using Principal Coordinate analysis (PCoA). The first two principal coordinate axes, which together explain 46.62% of variation, are shown. Each dot represents one taxonomic profile, with lines connecting profiles from the same sample. Colors represent developmental stages and shape represent profiling methods.