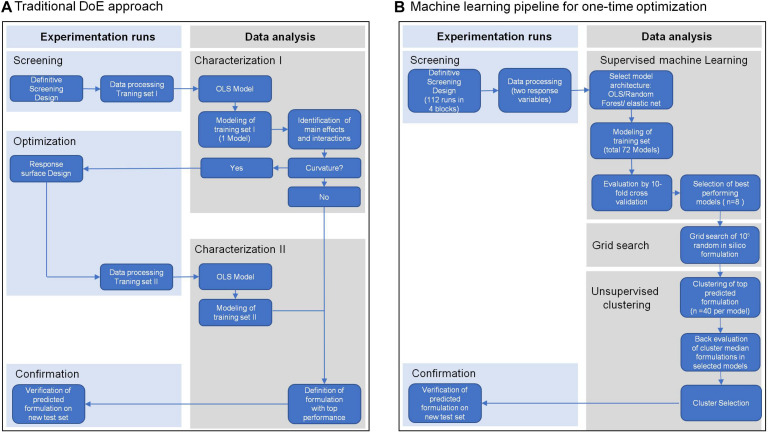
FIGURE 1.
Overview of the traditional Design of Experiment (DoE) approach (A) and of a machine learning pipeline (B). Both approaches include a screening step for data collection, in this case from expansion of T cells from four different healthy donors in a definitive screening design. Traditional DoE uses interpretable model architectures across all donors such as ordinary least squares regression (OLS) to select significant features (Characterization I). A second experimental step is applied to screen for optimal parameter levels (Optimization II). In a second modeling step using OLS the optimal parameter levels are determined for an optimal media formulation (Characterization II) that is experimentally confirmed (Confirmation). The machine learning pipeline (B) uses competitive machine learning algorithms to generate complex models for every response variable in every donor, which allow high prediction accuracy but are less interpretable (Supervised machine learning). After cross-validation these models are used to select the top 40 media formulations for every donor and every response variable from a random set of 105 in silico media formulations (Grid search). The top 40 formulations of all donors and responses are clustered by formulation similarity and a cluster formulation was defined by the median component level of all formulations in a cluster (Unsupervised Clustering). Back evaluation of cluster formulations in the donor models for every response allows selection of the media formulation with the best response across all donors and responses, which again is experimentally confirmed (Confirmation).