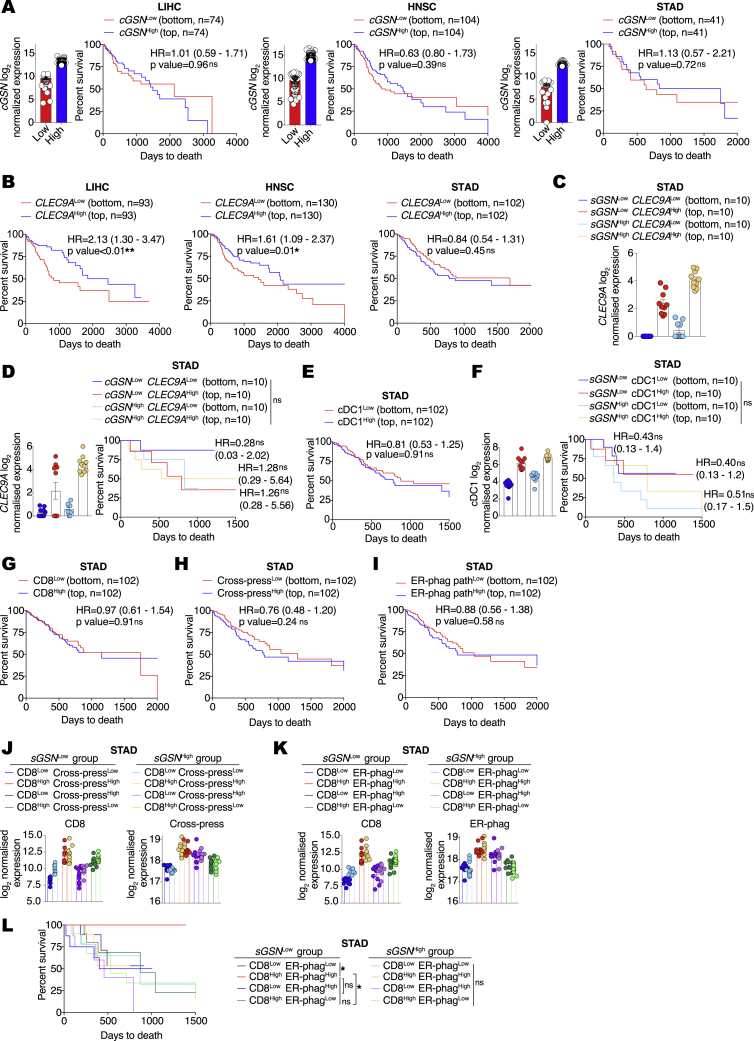
Figure S5.
Gene expression in human biopsies of LIHC, HNSC, and STAD tumors and association with patient survival, related to Figure 6
(A) Prognostic value of cytoplasmic gelslolin (cGSN) transcript levels for overall survival comparing samples with lowest (cGSNLow) and highest (cGSNHigh) expression in the indicated TCGA datasets. Liver hepatocellular carcinoma (LIH), bottom (n = 74) and top (n = 74) 20% of patient cohort. Head and neck squamous cell carcinoma (HNSC), bottom (n = 104) and top (n = 104) 20% of patient cohort. Stomach adenocarcinoma (STAD), bottom (n = 41) and top (n = 41) 10% of patient cohort.
(B) Prognostic value of CLEC9A expression for cancer patient overall survival comparing top and bottom quartiles in the indicated TCGA datasets.
(C) Transcript levels of CLEC9A expression comparing top and bottom quartiles of sGSNLow and sGSNHigh subgroups in the indicated TCGA datasets.
(D) Prognostic value of CLEC9A transcript levels expression for cancer patient overall survival comparing top and bottom quartiles of cGSNLow and cGSNHigh subgroups in the indicated TCGA dataset.
(E) Prognostic value of cDC1 gene signature expression for cancer patient overall survival comparing top and bottom quartiles in the indicated TCGA dataset.
(F) Prognostic value of cDC1 gene signature expression for cancer patient overall survival comparing top and bottom quartiles of sGSNLow and sGSNHigh subgroups in the indicated TCGA dataset.
(G–I) Prognostic value of (G) CD8 gene signature, (H) antigen processing and cross-presentation gene signature, (I) ER phagosome pathway gene signature expression for cancer patient overall survival comparing top and bottom quartiles in the indicated TCGA datasets.
(J) Transcript levels of CD8 and antigen processing and cross-presentation gene signatures comparing quartiles within sGSNLow and sGSNHigh subgroups in the indicated TCGA dataset.
(K and L) Transcript levels and synergistic prognostic value of CD8 and ER-phagosome pathway gene signatures comparing quartiles within sGSNLow and sGSN subgroups in the indicated TCGA dataset.
In (C, D, F, J, K, L) for cGSN and sGSN segregation between the highest and lowest expressors the same cut-off was used as in (A) for the indicated TCGA dataset. In (A, C, D, F, J, K) data are presented as mean of log2 normalized expression ± SEM Survival (Kaplan-Meier) curves in (A, B, D-F, G-I, L) were compared using Log-rank (Mantel-Cox) test. Hazard ratios (HR) with 95% confidence interval showed in brackets have been calculated in (A, B, E and G-I) as a ratio of Low expressed transcript /High expressed transcript group, in (D) as a ratio of each group / cGSNHighCLEC9ALow and in (F) as a ratio of each group / sGSNHighcDC1Low. ∗p ≤ 0.05, ∗∗p < 0.01, ns, not significant.