Highlights
-
•
The venom protein-protein interactions in snake venom remain largely unknown.
-
•
Y2H coupled with MD simulations was used to detect venom protein interactions.
-
•
Venom PLA2s interact with themselves and Lys49 PLA2 interacts with venom CRISP.
Keywords: Crotalus atrox, Venom protein, Protein-protein interaction, Yeast two-hybrid, Molecular dynamics simulation
Abstract
Proteins and peptides are major components of snake venom. Venom protein transcriptomes and proteomes of many snake species have been reported; however, snake venom complexity (i.e., the venom protein-protein interactions, PPIs) remains largely unknown. To detect the venom protein interactions, we used the most common snake venom component, phospholipase A2s (PLA2s) as a “bait” to identify the interactions between PLA2s and 14 of the most common proteins in Western diamondback rattlesnake (Crotalus atrox) venom by using yeast two-hybrid (Y2H) analysis, a technique used to detect PPIs. As a result, we identified PLA2s interacting with themselves, and lysing-49 PLA2 (Lys49 PLA2) interacting with venom cysteine-rich secretory protein (CRISP). To reveal the complex structure of Lys49 PLA2-CRISP interaction at the structural level, we first built the three-dimensional (3D) structures of Lys49 PLA2 and CRISP by a widely used computational program-MODELLER. The binding modes of Lys49 PLA2-CRISP interaction were then predicted through three different docking programs including ClusPro, ZDOCK and HADDOCK. Furthermore, the most likely complex structure of Lys49 PLA2-CRISP was inferred by molecular dynamic (MD) simulations with GROMACS software. The techniques used and results obtained from this study strengthen the understanding of snake venom protein interactions and pave the way for the study of animal venom complexity.
1. Introduction
Snakebite remains a major public health problem (Kasturiratne et al., 2008), particularly in impoverished rural communities (Gutiérrez et al., 2017). The World Health Organization therefore recognized snakebite as a high priority neglected tropical disease (WHO, 2018). The venom proteome and transcriptome of many snake species have been reported, such as European viper (Leonardi et al., 2019) and Sidewinder Rattlesnakes (Hofmann et al., 2018). Recently, Suryamohan et al. (2020) completed transcriptome and genome sequencing of Indian cobra. Moreover, the pathophysiological effects of many individual snake venom components have been characterized. However, snake venom is a complex mixture of proteins and peptides that are stored in the gland lumen to exert a wide range of toxic actions during envenomation; therefore, we hypothesize that venom proteins interact with each other to make a cocktail of proteins and peptides and synergistically exert their toxic effects. To our knowledge, in addition to the extensively reported lysing-49 PLA2s (Lys49 PLA2) homodimers (Almeida et al., 2016, Salvador et al., 2019), aspartic acid-49 PLA2(Asp49) homodimers (Corrêa et al., 2008) and Lys49-Asp49 heterodimer (Mora-Obando et al., 2014), little is known about the venom protein-protein interactions (PPIs), particularly the non-homologous venom protein interactions. Understanding the snake venom PPIs is critically important for deciphering the synergistic actions which exert various pathophysiological effects of snakebites.
Western diamondback rattlesnake (Crotalus atrox) is likely responsible for most snakebite fatalities in northern Mexico, and the second greatest number in the U.S.A. after Eastern diamondback rattlesnake (Crotalus adamanteus) (Campbell and Lamar, 2004). The proteome (Calvete et al., 2009) and incomplete transcriptome (Rokyta et al., 2011, Jia et al., 2020) of C. atrox venom protein have been reported; however, its venom PPIs remain poorly understood. The lack of a deep understanding of how C. atrox venom proteins interact is an important roadblock in advancing efforts to uncover the corresponding mechanisms of the synergistic effects for developing antivenom to treat snakebites. In our previous work (Jia et al., 2020), we identified 14 protein transcripts from the venom of C. atrox by using reverse transcription polymerase chain reaction (RT-PCR) and modeled their three-dimensional (3D) structures using predicted amino acid sequences and computational approach (MODELLER). These 14 transcripts are particularly important for us to further understand the complexity of C. atrox venom as well as to uncover the toxicity of each venom component by producing recombinant venom proteins. Therefore, to advance our knowledge about the venom complexity of C. atrox, in this study we attempted to detect the venom PPIs by using these 14 venom protein transcripts including PLA2s and cysteine-rich secretory protein (CRISP) transcripts. Snake venom PLA2, the most extensively studied venom components, is a major protein in most snake venoms. Snake venom PLA2s were classified into Group I (Elapidae) or Group II (Viperidae) of PLA2 superfamily that is composed of 16 Groups (Dennis et al., 2011). Group II snake venom PLA2s were further subdivided into at least two subgroups: the catalytically active Asp49-PLA2s which have an aspartic acid residue at the position 49, and catalytically inactive lys49-PLA2 homologue (or PLA2-like myotoxins) that the aspartic acid residue at the position 49 is replaced by Lysine (Renetseder et al., 1985, Maraganore and Heinrikson, 1986). In addition to the primary catalytic function, snake venom PLA2s display an array of pathophysiological effects including neurotoxicity, myotoxicity, cardiotoxicity, hemolytic activity, hypotensive activity, anticoagulant activity, etc. (Reviewed in Kini (2003)). CRISPs, single chain polypeptides with molecular weights of ~20–30 kDa, were found in mammalian epididymis and the immune system, and then in many snake venoms (Yamazaki and Morita, 2004). The overall structure of CRISP family proteins is well-conserved, consisting of an N-terminal domain (PR-1), and a conserved divalent metal-ion binding site that connected PR-1 to C-terminal domain (CR) (Shiol et al., 2019). The physiological functions of mammalian CRISPs, such as CRISP-1, -2 and -3, are associated with reproduction, cancer, and immune responses, while snake venom CRISPs inhibit ion channels (reviewed in Tadokoro et al. (2020)).
There are many molecular biology approaches that can be used to detect venom PPIs (Ho et al., 2002, Jares-Erijman and Jovin, 2006, Michnick et al., 2010). Among them, yeast two-hybrid (Y2H) analysis, since its inception (Fields and Song, 1989), has been extensively used to detect the physical interactions between any two proteins in different species. More importantly, Y2H analysis is amenable to being used in a high throughput setting, allowing a protein of interest to be tested for interactions with many proteins (Galletta and Rusan, 2015). For example, PPIs in yeast (Yu et al., 2008), nematode (Simonis et al., 2009), plant (Trigg et al., 2017, Marshall et al., 2019), human (Rolland et al., 2014) and bacterium (Rajagopala et al., 2014) were identified by high throughput Y2H analysis. By using Y2H analysis coupled with various computational programs including template-based modeling – MODELLER, protein-protein docking and molecular dynamics (MD) simulations, here we report the most common snake venom protein – phospholipase A2s (PLA2s) interacting with themselves and Lys49 PLA2 interacting with venom CRISP, as well as the most likely conformation of Lys49 PLA2-CRISP.
2. Materials and methods
2.1. Venom protein transcripts
Fourteen of the most common venom protein transcripts including Asp49 PLA2, Lys49 PLA2, snake venom metalloproteinase I (SVMPI), snake venom metalloproteinase II (SVMPII), snake venom metalloproteinase III (SVMPIII), snake venom serine proteinase (SVSP), L-amino acid oxidase (LAAO), C-type lectin, crotamine, three-finger toxin (3FTx), vespryn, CRISP, epidermal growth factor-like domain protein (EGF) and phospholipase B (PLB) used in this study, were obtained from our previous results (Jia et al., 2020). All translated amino acid sequences of these protein transcripts are available in our previous publication (Jia et al., 2020). All chemicals were purchased from Sigma-Aldrich (St. Louis, MO, USA).
2.2. Yeast two-hybrid analysis
Y2H activation and binding domain vectors (pAD and pBD) were purchased from Takara Bio USA, Inc (Mountain View, CA, USA). Polymerase Chain reactions (PCR) were carried out using cloned transcripts (Jia et al., 2020) as templates and Phusion High-Fidelity DNA Polymerase purchased from ThermoFisher Scientific (Waltham, MA, USA) by standard molecular biology methods (Table 1. Primer sequences). The PCR products of 14 mature transcripts were individually purified from agarose gel. Each transcript was ligated into EcoRI and BamHI sites of pAD vector as “prey” by using either restriction enzyme based-method or homologous recombination-based approach if enzymatic sites were present in the transcript sequence (such as EGF and SVSP). Using the same enzymatic method, transcripts for Lys49 PLA2, Asp49 PLA2 and CRISP were cloned into EcoRI and BamHI sites of pBD vector as “bait”. Empty vectors (pAD, pBD) served as negative controls. The conjunctions of recombinant DNAs were confirmed by Sanger sequencing, and the confirmed “bait” and “prey” constructs were co-transformed into yeast (Saccharomyces cerevisiae) strain (Y2HGold) cells (Takara Bio USA, Inc) based on the Takara instruction manual. The co-transformed cells were subsequently spread on double dropout (DDO, without tryptophan and leucine) medium and incubated at 30 °C until the colonies grew (~2 days) for confirming the successful pairwise transformation. Plasmid DNAs were extracted from co-transformed cells to verify the presence of both “bait” and “prey”. Verified single co-transformed colonies were diluted to optical density OD600 = 1 in distilled water. Equal amounts (5 μl) of each colony were spotted on DDO and quadruple dropout (QDO) medium (without tryptophan, leucine, histidine, and adenine). QDO plates were incubated at 30 °C for 55 h to detect PPIs and photographed. X-alpha-Gal (40 μg/ml) was added in QDO medium for ‘reciprocal’ Y2H (rY2H) (swapping “bait” and “prey” venom proteins between pAD and pBD vectors). At least 3 different colonies from each co-transformation were spotted on both DDO and QDO mediums to validate the reproducibility of results.
Table 1.
PCR primer sequences.
| Transcript | Primer pair | Primer sequences (5′–3′) |
|---|---|---|
| Lys49 PLA2 | Lys49-EcoRI F Lys49-BamH I R |
GGTGAATTCAGCCTGGTCGAATTGGGGAA GGTGGATCCTCATTAGCATGTATCTGGCTTCTT |
| Asp49 PLA2 | Asp49-EcoRI F Asp49-BamH I R |
GGTGAATTCAACCTGCTGCAATTCAACAA GGTGGATCCTCATTAGCATTTCTCTGAAGGGT |
| SVMP I | MPI-EcoRI F MPI-BamHI R |
GGTGAATTCGTGAATGATTATGAAGTA GGTGGATCCTCATTAAGCCTCCAAAAGTTCATT |
| SVMP II | MPII-EcoRI F MPII-BamHI R |
GGTGAATTCATAATCCTGGAATCTGGGA GGTGGATCCTCATTAGCCATAGAGGCCATTT |
| SVMP III | MPIII-EcoRI F MPIII-BamHI R |
GGTGAATTCATAATCCTGGAATCTGGGA GGTGGATCCTCACTAGTAGGCTGTAGCCA |
| C-type lectin | Lect-EcoRI F Lect-BamHI R |
GGTGAATTCGATTGTCCCTCTGGTT GGTGGATCCTCACTATGCCTTGCAGACGA |
| 3FTx | 3FTx-EcoRI F 3FTx-BamHI R |
GGTGAATTCCTGGAATGTGAAGCATGCAA GGTGGATCCTCATTAAGCGTTGCACAGGTT |
| CRISP | Cri-EcoRI F Cri-BamHI R |
GGTGAATTCAGTGTTGATTTTGATT GGTGGATCCTCACTATATTATTTTATTT |
| Vespryn | Ves-EcoRI F Ves-BamHI R |
GGTGAATTCGATGTGACGTTTGACTCAA GGTGGATCCTCATTAAAGAGTTGTGAGT |
| Crotamine | Cro-EcoRI F Cro-BamHI R |
GGTGAATTCCAATCACAGTGTGAACA GGTGGATCCTCATTATTTTCCAATTTTGCT |
| LAAO | LAAO-EcoRI F LAAO-BamHI R |
GGTGAATTCATGTCTTCTGTGACAGTT GGTGGATCCTCATTAAAATTCATTGTCAT |
| PLB | PLB-EcoRI F PLB-BamHI R |
GGTGAATTCGATATCCACTATGCTA GGTGGATCCTCATCACAGCACTGGTTTCA |
| EGF* | EGF-pBD F EGF-pBD R |
CATGGAGGCCGAATTCCTCTGGGCACCTCCGA GCAGGTCGACGGATCCAAAGTCTTCATGGGTA |
| EGF* | EGF-pAD F EGF-pAD R |
GGAGGCCAGTGAATTCCTCTGGGCACCTCCGA CGAGCTCGATGGATCCAAAGTCTTCATGGGTA |
| SVSP* | Sp-pBD F SP-pBD R |
CATGGAGGCCGAATTCGTCGTTGGAGGTGATGAA GCAGGTCGACGGATCCATGGGGGGCAGGTCGCAT |
| SVSP* | SP-pAD F SP-pAD R |
GGAGGCCAGTGAATTCGTCGTTGGAGGTGATGAA CGAGCTCGATGGATCCATGGGGGGCAGGTCGCAT |
*Homologous recombination-technique was used for both EGF and SVSP due to either EcoRI or BamHI site presenting in the open-reading frames.
2.3. Molecular dynamics simulation
Amino acid sequences of Lys49 PLA2 and CRISP were blasted against Protein Data Bank (PDB) (Berman et al., 2007). Crystal structures with PDB codes for 6CE2 and 1RC9 corresponding to Lys49 PLA2 and CRISP were selected as the templates for modeling the 3D structures of Lys49 PLA2 and CRISP by the most widely used template-based protein structure modeling software, MODELLER version 9.24 (Ŝali and Blundell, 1993, Webb and Ŝali, 2014), using the procedures detailed in previous work (Jia et al., 2020). But this time, we simulated 100 candidate models for each venom protein and selected the one with the lowest discrete optimized protein energy (DOPE) score for subsequent protein-protein docking programs. The modeled structures of Lys49PLA2 and CRISP were evaluated through the following criteria: 1) DOPE score (Webb and Ŝali, 2014), the lowest DOPE score model represents the more accurate protein model at its native conformation; 2) overall quality factor (ERRAT) (Colovos and Yeates, 1993), checking if the score is above the expected accuracy of 70% of residues for medium resolution structure; 3) Verify3D (Eisenberg et al., 1997), testing if more than 70% residues having compatibility between the 3D model and the amino acid sequence (1D); 4) Z-score (Wiederstein and Sippl, 2007), testing if the protein model predicted falls within range of high quality experimental structure with a similar size and shape; and 5) Ramachandran (φ/ψ) plot (Ramachandran et al., 1963), checking if interrogated phi and psi dihedral angles of more than 90% of C-alpha residues were within the protein model. All 3D structures of Lys49 PLA2, CRISP and Lys49 PLA2-CRISP were revealed by UCSF Chimera program (Pettersen et al., 2004). Further, three top listed docking programs including ZDOCK (Pierce et al., 2014), HADDOCK (van Zundert et al., 2016) and ClusPro (Kozakov et al., 2017) were used to predict the binding modes of Lys49 PLA2-CRISP interaction using default settings except HADDOCK by activation of random patches. The top docked modes from each docking program were selected for MD simulations by using GROMACS 2020 package (Hess et al., 2008). The CHARMM36 force field (Best et al., 2012) and the standard TIP3P water model (Jorgensen et al., 1983) were chosen for all MD simulations. 1) Each complex was embedded in the center of a dodecahedron water box with a minimum distance from the complex to the box boundary of 10 Å. 2) The complex was solved with water modeled by the TIP3P force field. 3) The system was further neutralized by proper NaCl solution. 4) Energy minimization was carried out for each complex using a steepest-descent integrator to reach negative potential energy and the maximum force for less than 1,000 kJ/(mol.nm) on any atom. 5) To relax the protein complex, the equilibration was performed with positional restraints on all heavy atoms of complex under two ensembles, NVT (constant number of particles, volume and temperature) under constant temperature at 300 K for 100 picoseconds (ps), and NPT (constant number of particles, pressure, temperature) under constant pressure at 1 bar for 100 ps. 6) The production MD simulation was conducted for 1 nanosecond (ns) for initial screening, then 10 ns for further confirmation of structure stability by using the same settings as the previous equilibration but without restraints. The complex structure stability of Lys49 PLA2 interacting with CRISP was assessed by measuring room-mean-square deviation (RMSD) for backbone atoms of each complex. The RMSDs were illustrated with R-program (Team, 2013).
3. Results
3.1. Venom Lys49 PLA2 interacts with CRISP
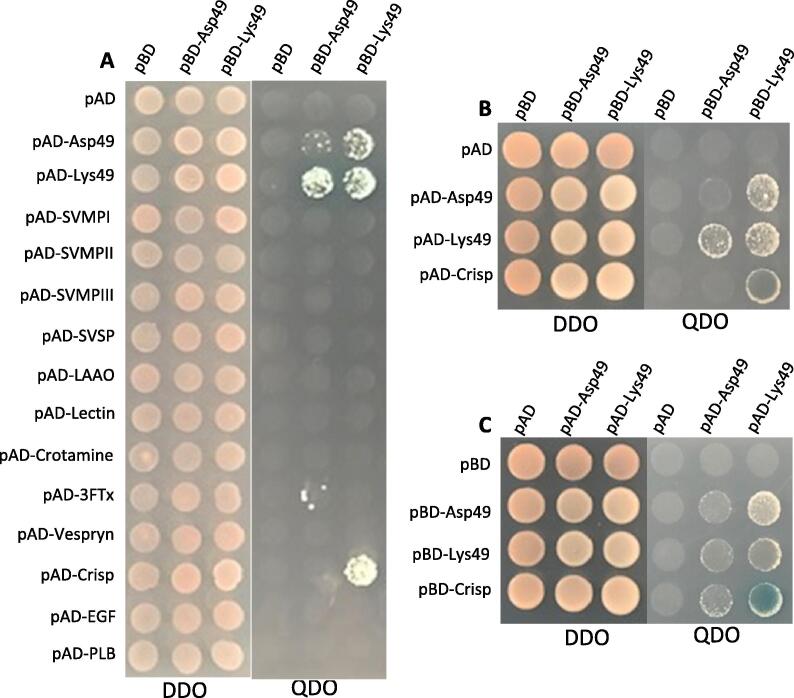
We screened C. atrox venom proteins encoded by 14 of the most common transcripts for detecting the venom PPIs using the most common venom component, PLA2 including Lys49 PLA2 and Asp49 PLA2 as “bait” by Y2H analysis. The screening results showed that PLA2s interact with themselves, which can be served as perfect internal positive controls in Y2H analysis because native venom PLA2 dimers have been extensively reported, including C. atrox Lys49 PLA2 homodimer (Brunie et al., 1985) and Asp49 PLA2 homodimer (Keith et al., 1981). Moreover, Lys49 PLA2 strongly interacts with venom CRISP, while Asp49 PLA2 weakly interacts with 3FTx (Fig. 1A). To eliminate the false positives, we deployed “rY2H” analysis by swapping “bait” and “prey” proteins between pAD and pBD vectors (i.e., pBD-Lys49 PLA2 × pAD-CRISP, and pBD-CRISP × pAD-Lys49 PLA2). Briefly, 1) we repeated above screening results but individually and with more stringent detection by adding X-alpha-Gal. The results are reproducible in that PLA2S form dimers and Lys49 PLA2 interacts with CRISP (Fig. 1B). 2) To further verify the genuine interaction between Lys49 PLA2s and CRISP, we switched Lys49 PLA2 to pAD and CRISP to pBD vectors (Fig. 1C), yielding the same results as in Fig. 1B except the ‘unilateral’ interaction between Asp49 PLA2 and CRISP (Fig. 1C). Empty vectors (pAD and pBD) served as negative controls showing no interactions with any venom proteins (Fig. 1 A, B and C).
Fig. 1.
Yeast two-hybrid analysis. Colony growth on DDO medium (without leucine and tryptophan) indicates the successful binary co-transformations, while colony growth on QDO medium (without histidine, leucine, tryptophan, and adenine) shows the protein-protein interactions (PPIs). A, using pBD-Asp49 PLA2 and pBD-Lys49 PLA2 as “bait” to detect the venom PPIs between PLA2s and 14 venom proteins in pAD vector (empty pBD and pAD served as negative control). B, C, the results of ‘reciprocal’ Y2H, swapping venom proteins from pBD (B) to pAD vector (C), show that PLA2s form dimers and Lys49 PLA2 interacts with CRISP on QDO/X-alpha-Gal medium.
3.2. The stable and possible binding mode of Lys49 PLA2-CRISP
Y2H technique can be used to identify PPIs, but it is unable to reveal the complex 3D structure of PPIs. To display the 3D structure of Lys49 PLA2-CRISP interaction, we first built the models for Lys49 PLA2 and CRISP based on the crystallographic structures of MjTX-I (PDB ID 6CE2) from Bothrops moojeni venom (Salvador et al., 2018) and Stecrisp (PDB ID 1RC9) from Trimeresurus Stejnegeri venom (Guo et al., 2004), respectively. The amino acid sequences of the models shared higher sequence identity (more than 70%) and overall sequence coverage of 100% to that of the templates (Table 2). The RMSD values after superimposition and aligning all atoms of models (Lys49 PLA2 and CRISP) to the templates (6CE2 and 1RC9) were 0.154 and 0.200 Å, respectively. This suggests very little deviation between carbon main chain atoms of models and templates, demonstrating that modeled structures are near to the native structures. To further evaluate the accuracy and reliability of modeled structures, we used various stereochemical parameters including ERRAT, vertify3D, Z-Score, and backbone φ/ψ angles. The results shown in Table 3 support that predicted models are of good quality and satisfied the quality checks. After choosing the validated models, we conducted the protein-protein docking, followed by MD simulations. The top complex structures of 11, 9 and 8 were generated by ClusPro, HADDOCK and ZDOCK docking programs, respectively. Subsequently, the great challenge is to identify the correct binding modes of Lys49 PLA2 and CRISP interaction. Fortunately, MD simulation is a well-established technique and can be used for identifying the possible correct binding mode by assessing the time-resolved motions and stability of protein-protein complex structures at atomic resolution (Radom et al., 2018, Sakano et al., 2016, Dror et al., 2012, Perilla et al., 2015). Therefore, we determined which complex structure forms the stable conformation by using MD simulations. All above 28 docked structures were run for 1000 ps (Fig. 2A) by using GROMACS. After MD simulations, the three docking poses, model3, cluster21 and complex6 with average complex backbone RMSDs of 0.229 ± 0.034 nm2, 0.245 ± 0.054 nm2 and 0.363 ± 0.058 nm2, respectively, show the most stable binding modes in each group (Fig. 2D, 2E, 2F). We further conducted MD simulations for these three top complex structures by extending time to 10 ns, resulting that model3 is the most stable complex structure and therefore is the most likely docking mode for Lys49-CRISP interaction, whereas complex6 and particularly cluster21 quickly shifted from initial mode at the time of 2 ns (Fig. 2G). During 10 ns simulations, the average RMSD of backbone atoms of the three systems (Lys49 PLA2, CRISP and model3) indicated that CRISP (0.21 ± 0.02) had very similar RMSD values compared to 0.23 ± 0.02 for model3, whereas Lys49 PLA2 had even lower RMSD (0.14 ± 0.03), suggesting that three systems show similar behavior (Fig. 2G). Additionally, there are 3 hydrogen bonds formatted between Lys49 PLA2 and CRISP within 3 Å in model3, and no inter-residue-hydrogen bonds formatted in cluster21 and complex6 (Fig. 2D, 2E, 2F).
Table 2.
BLASTP analysis of Lys49 PLA2 and CRISP and the selected templates.
| Models | Templates | Identity (%) | Similarity (%) | Cover (%) | Mas score | E-values |
|---|---|---|---|---|---|---|
| Lys49 PLA2 | 6CE2 | 72 | 80 | 100 | 176 | 4e−58 |
| CRISP | 1RC9 | 81 | 91 | 100 | 391 | 4e−140 |
Table 3.
Evaluation of Lys49 PLA2 and CRISP models.
| Models | DOPE | ERRAT (%) | Verify3D (%) | Z-score | φ/ψ-plot (%) |
|---|---|---|---|---|---|
| Lys49 PLA2 | −11141.5 | 82.3 | 87.6 | −5.0 | 91.4 |
| CRISP | −22896.4 | 86.6 | 88.2 | −5.7 | 92.8 |
DOPE (Discrete Optimized Protein Energy) scores were generated via many iterations by MODELLER (Webb et al., 2014) scripts; more negative DOPE score values tend to correlate with more native-like models. ERRAT scores over 80% indicate that only a few residues have an elevated error function when compared with similar experimental structures. Verify3D scores over 80% signify that the amino acids have compatibility between the 3D model and the amino acid sequence. Z-score is used to assess if the knowledge-based potential could recognize a native fold from the other alternatives (acceptable range from −12 to 12). Ramachandran plot is used to test if protein amino acid residues are in most favored phi (φ) and psi (ψ) dihedral angle region. ERRAT, Verify3D, Z-score and Ramachandran plot were calculated by PROCHECK (Laskowski et al., 1993).
Fig. 2.
Prediction of Lys49 PLA2-CRISP complex structure by molecular dynamics (MD) simulations. Room-mean-square deviation (RMSD) of backbone atoms from docked modes of (Lys49-CRISP) as a function of simulation time. A, B and C: During the simulation (1 ns), some complexes in each docking method (ClusPro, ZDOCK and HADDOCK) move away from the initial pose (the trajectory drift away from the initial structure), while the others remain close. The former could be identified as wrong docking models, and the latter as possible correct models (e.g., model3, cluster21, complex6). D, E and F: The 3D complex structures of model3, cluster21 and complex6 (CRISP in magenta, Lys49 PLA2 in blue, and hydrogen bonds denoted by black line in model3), visualized by UCFC Chimera. G: The comparison of structure stability of Lys49 PLA2, CRISP and three complexes in extended time (10 ns). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
4. Discussion
4.1. Venom protein interaction
Proteins and peptides constitute the complex mixture of snake venom and understanding the venom complexity is critical for developing antivenom for the treatment of snakebites. However, to the best of our knowledge, there is no systematic report of even a single venom complexity, partially due to the lack of high-throughput technique for detecting venom PPIs. Fortunately, since its inception, Y2H analysis has been extensively used to detect any PPIs. Certainly, like all the other approaches, there are some limitations of Y2H analysis. The major drawback of Y2H analysis is that it generates false positives. Many researchers utilized different approaches such as different Y2H systems (Caufield et al., 2012), pull-down analysis (Rimbault et al., 2019), size exclusion (Kirkwood et al., 2013, Busch et al., 2018), etc. as complementary approaches to validate the Y2H results. Among them, a widely used method is to increase the stringency of the interactions. In the present study, apart from using stringent interactions by adding X-alpha-Gal, we deployed “rY2H” method to validate the true interactions. We only consider the PPIs from “rY2H” results as the guanine interactions, e.g., interactions between Lys49 PLA2 and CRISP, as well as Lys49 PLA2 homodimer and PLA2 heterodimer (Fig. 1A, 1B, and 1C). Braun et al., 2009, Caufield et al., 2012 also successfully validated the true PPIs by swapping bait-prey proteins in Y2H systems. PPIs from ‘unilateral’ Y2H need to be further confirmed such as the interactions of Asp49 PLA2 and CRISP in Fig. 1C but not in Fig. 1B, as well as Asp49 PLA2-3FTx interaction in Fig. 1A.
PLA2 enzymes are one of the most common proteins in most snake venoms and are responsible for diverse toxicities including neurotoxicity, myotoxicity, blood coagulation, etc. (Kini, 2003, Lomonte et al., 2003). We predicted that PLA2 interacting with other venom proteins plays a key role in pathophysiological effects. Surprisingly, our findings demonstrated that venom PLA2 only interacts with few venom proteins such as PLA2 itself, CRISP, and possibly with 3FTx. This result implies that venom toxins probably either execute toxic effects individually, or transiently interact with other molecules to exhibit synergistic activity during envenomation. The most common function of snake venom CRISP is to inhibit ion channels, but no snake venom CRISP so far have proved lethal to mammals (reviewed in Tadokoro et al. (2020)), whereas Lys49 PLA2s were extensively reported that they possess myotoxic activity; thus, we speculate that the interaction of Lys49 PLA2 and CRIDP probably produce more potent myotoxicity or strong binding capacity to various target molecules such as ion channels to affect cellular signaling. Our findings are in good agreement with prior conclusion that most snake venom toxins exhibit their pharmacological activities on their own; however, some proteins form covalent/non-covalent complexes with other proteins to exhibit more potent pharmacological activities (reviewed in Doley and Kini (2009)). It appears that snake venom proteins preferentially form homodimers or heterodimers with different members of the same venom protein family, and they scarcely interact with non-homologous venom proteins. Understanding this principle is critical for the development of universal antivenom or complement methods to treat snakebite, the neglected tropical disease that results in high mortality (138,000 deaths per annum) and morbidity (~400, 000 cases per annum) (Gutiérrez et al., 2017).
4.2. Complex structure of venom protein interactions
While the successful detection of venom PPIs is important, the study of venom protein interactions at the structural level plays a crucial role in structure-based antivenom development because the pathophysiological effects of venom proteins or protein complexes are known to be closely related to their 3D structures. To predict the complex structure of Lys49 PLA2-CRISP interaction, the accuracy and reliability of modeled individual structures of Lys49 PLA2 and CRISP were critically important for following docking and MD simulations. Both modeled structures of Lys49 PLA2 and CRISP are close to the native structures with less than 0.2 Å RMSD between templates and modeled structures. Furthermore, the various structure parameters indicate that the modeled structures of Lys49 and CRISP are close to the crystal experimental structures (Table 3). Subsequently, based on Critical Assessment of protein Structure Prediction (CASP)-Critical Assessment of PRediction of Interactions (CAPRI) (Lensink et al., 2017, Vangone et al., 2017, Kurkcuoglu and Bonvin, 2019, Agrawal et al., 2019), we employed the three top ranked protein-protein docking methods (ClusPro, ZDOCK and HADDOCK) for predicting the complex structure of Lys49 PLA2-CRISP interaction. To further identify the most likely docking mode of Lys49 PLA2-CRISP interactions, we performed the MD simulations and the results revealed that the model3 (Fig. 2D and G) shows stable complex structure in extended time (10 ns) and exhibits similar stable behavior with Lys49 PLA2 and CRISP (Fig. 2G) when comparing with cluster21 and complex6. MD simulation can evaluate the thermodynamic stability of protein complex structures in time-resolved motions, and the highly stable complex structures should represent the most likely biological form of functional structure (Krissinel and Henrick, 2007). In addition, MD simulation has been tested for several systems to identify the correctly docked modes (Dror et al., 2011, Shan et al., 2011, Buch et al., 2011), and it is commonly accepted that the correctly docked conformations appear to be thermodynamically stable (Quezada et al., 2017, Radom et al., 2018, Bhakat et al., 2018).
The study of venom protein interactions at the structural level plays a crucial role in the investigation of toxic systems and the development of antivenom. However, the current situation in animal venom research is that there are numerous venom protein transcripts and even protein sequences available in various databases. For example, there are 10 transcript sequences of Crotalus CRISP deposited in NCBI GenBank, but only one crystal structure of Crotalus CRISP is available in PDB (accessed in January 2021). Therefore, there is a large gap between the number of available venom protein sequences and their experimentally solved crystal structures. Developing crystal structures of venom proteins is a time consuming and tedious laboratory process. To fill out this gap, computational modeling of the 3D structure of complexes from their protein sequences play a central role in structural venom research. From our current study, we believe that the combination of experimental (Y2H) and computational (modeling) approaches appears promising for not only detections of venom PPIs, but also for dissection of venom complexity at the structural level.
5. Conclusions
Understanding the venom protein interactions is essential for deciphering venom complexity and designing specific chemical modifications to develop new reagents and therapeutics for the treatment of snakebites. We detected the venom protein-protein interactions of C. atrox and the results showed that venom PLA2s interact with themselves and Lys49 PLA2 interacts with CRISP. The thermodynamically stable conformations of Lys49-CRISP were further predicted by MD simulations. Based on the above and our other preliminary results (data not shown), we propose that, although snake venom is a cocktail of proteins and other molecules, most venom proteins individually exert their pathophysiological effects or are coupled with small molecules such as peptides and chemical compounds. Future studies will entail testing the synergistic activities of Lys49-CRISP complex.
CRediT authorship contribution statement
Paulina Kowalski: Methodology, Formal analysis, Investigation, review & editing. Ivan Lopez: Data curation, software, review & editing. Ying Jia: Supervision, Validation, Writing - original draft, Writing - review & editing.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Authors are grateful for the support from the University of Texas Rio Grande Valley (UTRGV) Engaged Scholar and Artist Awards (ESAA) and UTRGV High Scholars Program (HSP).
References
- Agrawal P., Singh H., Srivastava H.K., Singh S., Kishore G., Raghava G.P.S. Benchmarking of different molecular docking methods for protein-peptide docking. BMC Bioinf. 2019;19(Suppl. 13):426. doi: 10.1186/s12859-018-2449-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida J.R., Lancellotti M., Soares A.M., Calderon L.A., Ramírez D., González W., Marangoni S., Da Silva S.L. CoaTx-II, a new dimeric Lys49 phospholipase A2 from Crotalus oreganus abyssus snake venom with bactericidal potential: insights into its structure and biological roles. Toxicon. 2016;120:147–158. doi: 10.1016/j.toxicon.2016.08.007. [DOI] [PubMed] [Google Scholar]
- Berman H., Henrick K., Nakamura H., Markley J.L. The worldwide protein data bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Best R.B., Zhu X., Shim J., Lopes P.E.M., Mittal J., Feig M., MacKerell A., Jr. Optimization of the additive CHARMM all atom protein force field targeting improved sampling of the backbone phi, psi and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 2012;8:3257–3273. doi: 10.1021/ct300400x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakat S., Åberg E., Söderhjelm P. Prediction of binding poses to FXR using multitargeted docking combined with molecular dynamics and enhanced exampling. J. Comput. Aided Mol. Des. 2018;32:59–73. doi: 10.1007/s10822-017-0074-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun P., Tasan M., Dreze M., Barrios-Rodiles M., Lemmens I., Yu H. An experimentally derived confidence score for binary protein-protein interactions. Nat. Methods. 2009;6:91–97. doi: 10.1038/nmeth.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunie S., Bolin J., Gewirth D., Sigler P.B. The refined crystal structure of dimeric phospholipase A2 at 2.5 Å. J. Biol. Chem. 1985;260:9742–9749. [PubMed] [Google Scholar]
- Buch I., Giogino T., de Fabritiis G. Complete reconstruction of an enzyme-inhibitor binding process by molecular dynamics simulations. Proc. Natl. Acad. Sci. U.S.A. 2011;108:10184–10189. doi: 10.1073/pnas.1103547108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch F.M., Sahasrabuddhe A., vanAernum Z., Rivera B., Wysocki V.H. Analysis of proteins and protein interactions by size exclusion chromatography-high resolution mass spectrometry. FASER J. 2018;31(Supplement 926):9. [Google Scholar]
- Calvete J., Fasoli E., Sanz L., Boschetti E., Righetti P.G. Exploring the venom proteome of the western diamondback rattlesnake, Crotalus atrox, via snake venomics and combinatorial peptide ligand library approaches. J. Proteome Res. 2009;8:3055–3067. doi: 10.1021/pr900249q. [DOI] [PubMed] [Google Scholar]
- Campbell J.A., Lamar W.W. Comstock Publishing Associates; Ithaca, NY: 2004. The Venomous Reptiles of the Western Hemisphere. [Google Scholar]
- Caufield J.H., Sakhawalkar N., Uetz P. A comparison and optimization of yeast two-hybrid systems. Methods. 2012;58:317–324. doi: 10.1016/j.ymeth.2012.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colovos C., Yeates T.O. Verification of protein structure: patterns of nonbonded atomic interactions. Protein Sci. 1993;2:1511–1519. doi: 10.1002/pro.5560020916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrêa L.C., Marchi-Salvador D.P., Cintra A.C.O., Sampaio S.V., Soares A.M., Fontes M.R.M. Crystal structure of a myotoxic Asp49-phospholipase A2 with low catalytic activity: insights into Ca2+-independent catalytic mechanism. Biochim. Biophys. Acta. 2008;1784:591–599. doi: 10.1016/j.bbapap.2008.01.007. [DOI] [PubMed] [Google Scholar]
- Dennis E.A., Cao J., Hsu Y.H., Magrioti V., Kokotos G. Phospholipase A2 enzymes: physical structure, biological function, disease implication, chemical inhibition, and therapeutic intervention. Chem. Rev. 2011;111:6130–6185. doi: 10.1021/cr200085w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doley R., Kini R.M. Protein complexes in snake venom. Cell. Mol. Life Sci. 2009;66:2851–2871. doi: 10.1007/s00018-009-0050-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror R.O., Pan A.C., Arlow D.H., Borhani D.W., Maragakis P., Shan Y., Xu H., Shaw D.E. Pathway and mechanism of drug binding to G-protein-coupled receptors. Proc. Natl. Acad. Sci. U.S.A. 2011;108:13118–13123. doi: 10.1073/pnas.1104614108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror R.O., Dirks R.M., Grossman J.P., Xu H., Shaw D.E. Biomolecular simulation: a computational microscope for molecular biology. Annu. Rev. Biophys. 2012;41:429–452. doi: 10.1146/annurev-biophys-042910-155245. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Lüthy R., Bowie J.U. VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol. 1997;277:396–404. doi: 10.1016/s0076-6879(97)77022-8. [DOI] [PubMed] [Google Scholar]
- Fields S., Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- Galletta B.J., Rusan N.M. A yeast two-hybrid approach for probing protein-protein interactions at the centrosome. Methods Cell Biol. 2015;129:251–277. doi: 10.1016/bs.mcb.2015.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Teng M., Niu L., Liu Q., Huang Q., Hao Q. Crystal structure of cysteine-rich secretory protein stecrisp reveals the cysteine-rich domain has a K+-channel inhibitor-like fold. J. Biol. Chem. 2004;280:12405–12412. doi: 10.1074/jbc.M413566200. [DOI] [PubMed] [Google Scholar]
- Gutiérrez J.M., Calvete J.J., Habib A.G., Harrison R.A., Williams D.J., Warrell D.A. Snakebite envenoming. Nat. Rev. Dis. Primers. 2017;3:17079. doi: 10.1038/nrdp.2017.79. [DOI] [PubMed] [Google Scholar]
- Hess B., Kutzner C., van der Spoel D., Lindahl E. GROMACS 4: algorithms for highly efficient, load-balanced, and scalable molecular simulation. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Ho Y., Gruhler A., Heilbut A., Bader G.D., Moore I. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. [DOI] [PubMed] [Google Scholar]
- Hofmann E.P., Rautsaw R.M., Strickland J.L., Holding M.L., Hogan M.P., Mason A.J., Rokyta D.R., Parkinson C.L. Comparative venom-gland transcriptomics and venom proteomics of four Sidewinder Rattlesnake (Crotalus cerastes) lineages reveal little differential expression despite individual variation. Sci. Rep. 2018;8:15534. doi: 10.1038/s41598-018-33943-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jares-Erijman E.A., Jovin T.M. Imaging molecular interactions in living cells by FRET microscopy. Curr. Opin. Chem. Biol. 2006;10:409–416. doi: 10.1016/j.cbpa.2006.08.021. [DOI] [PubMed] [Google Scholar]
- Jia Y., Lopez I., Kowalski P. Toxin transcripts in Crotalus atrox and in silico structures of toxins. J. Venom Res. 2020;10:18–22. [PMC free article] [PubMed] [Google Scholar]
- Jorgensen W.L., Chandrasekhar J., Madura J.D., Impey R.W., Klein M.L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 1983;79:926. [Google Scholar]
- Kasturiratne A., Wickremasinghe A.R., De Silva N., Gunawardena N.K., Pathmeswaran A., Premaratna R. The global burden of snakebite: a literature analysis and modelling based on regional estimates of envenoming and deaths. PLoS Med. 2008;5 doi: 10.1371/journal.pmed.0050218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keith C., Feldman D.S., Deganello S., Glick J., Ward K.B., Jones E.O., Sigler P.B. The 2.5 Å crystal structure of a dimeric phospholipase A2 from the venom of Crotalus atrox. J. Biol. Chem. 1981;256:8602–8607. [PubMed] [Google Scholar]
- Kini R.M. Excitement ahead: structure, function and mechanism of snake venom phospholipase A2 enzymes. Toxicon. 2003;42:827–840. doi: 10.1016/j.toxicon.2003.11.002. [DOI] [PubMed] [Google Scholar]
- Kirkwood K.J., Ahmad Y., Larance M., Lamond A.I. Characterization of native protein complexes and protein isoform variation using size-fractionation-based quantitative proteomics. Mol. Cell. Proteomics. 2013;12:3851–3873. doi: 10.1074/mcp.M113.032367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozakov D., Hall D.R., Xia B., Porter K.A., Padhorny D., Yueh C., Beglov D., Vajda S. The ClusPro web server for protein-protein docking. Nat. Protoc. 2017;12:255–278. doi: 10.1038/nprot.2016.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurkcuoglu Z., Bonvin A.M.J.J. Pre- and post-docking sampling of conformational changes using ClusENM and HADDOCK for protein-protein and protein-DNA systems. Proteins. 2019;88:292–306. doi: 10.1002/prot.25802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krissinel E., Henrick K. Inference of macromolecular assemblies from crystalline state. JMB. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- Laskowski R.A., MacArthur M.W., Moss D.S., Thornton J.M. PROCHECK: a program to check the stereochemical quality of protein structure. J. Appl. Crystallogr. 1993;26:283–291. [Google Scholar]
- Lensink M.F., Velankar S., Wodak S.J. Modeling protein-protein and protein-peptide complexes: CAPRI. Proteins. 2017;85:359–377. doi: 10.1002/prot.25215. [DOI] [PubMed] [Google Scholar]
- Leonardi A., Sajevic T., Pungerčar J., Križaj I. Comprehensive study of the proteome and transcriptome of the venom of the most venomous european viper: discovery of a new subclass of ancestral snake venom metalloproteinase precursor-derived proteins. J. Proteome Res. 2019;18:2287–2309. doi: 10.1021/acs.jproteome.9b00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lomonte B., Angulo Y., Calderόn L. An overview of lysine-49 phospholipase A2 myotoxins from crotalid snake venoms and their structural determinants of myotoxic action. Toxicon. 2003;42:885–901. doi: 10.1016/j.toxicon.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Maraganore J.M., Heinrikson R.L. The lysine-49 phospholipase A2 from the venom of Agkistrodon piscivorus piscivorus. Relation of structure and function to other phospholipases A2. J. Biol. Chem. 1986;261:4797–4804. [PubMed] [Google Scholar]
- Marshall R.S., Hua Z., Mali S., McLoughlin F., Vierstra R.D. ATG8-binding UIM proteins define a new class of autophagy adaptors and receptors. Cell. 2019;177:766–781. doi: 10.1016/j.cell.2019.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michnick S.W., Ear P.H., Landry C., Malleshaiah M.K., Messier V. A toolkit of protein-fragment complementation assays for studying and dissecting large-scale and dynamic protein-protein interactions in living cells. Methods Enzymol. 2010;470:335–368. doi: 10.1016/S0076-6879(10)70014-8. [DOI] [PubMed] [Google Scholar]
- Mora-Obando D., Fernández J., Montecucco C., Gutiérrez J.M., Lomonte B. Synergism between basic Asp49 and Lys49 phospholipase A2 myotoxins of viperid snake venom in vitro and in vivo. PLoS One. 2014;9 doi: 10.1371/journal.pone.0109846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perilla J.R., Goh B.C., Cassidy C.K., Liu B., Bernardi R.C., Rudack T., Yu H., Wu Z., Schulten K. Molecular dynamics simulations of large macromolecular complexes. Curr. Opin. Struct. Biol. 2015;31:64–74. doi: 10.1016/j.sbi.2015.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera a visualization system for exploratory 729 research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pierce B.G., Wiehe K., Hwang H., Kim B.H., Vreven T., Weng Z. ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers. Bioinformatics. 2014;30:1771–1773. doi: 10.1093/bioinformatics/btu097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quezada A.G., Díaz-Salazara A.J., Cabrera N., Pérez-Montfort R., Pineiro A., Costas M. Interplay between protein thermal flexibility and kinetic stability. Structure. 2017;25:167–179. doi: 10.1016/j.str.2016.11.018. [DOI] [PubMed] [Google Scholar]
- Radom F., Plückthun A., Paci E. Assessment of ab initio models of protein complexes by molecular dynamics. PLoS Comput. Biol. 2018;14 doi: 10.1371/journal.pcbi.1006182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rajagopala S.V., Sikorski P., Kumar A., Mosca R., Vlasblom J., Arnold R., Franca-Koh J., Pakala S.B., Phanse S. The Binary protein-protein interaction landscape of Escherichia coli. Nat. Biotechnol. 2014;32:285. doi: 10.1038/nbt.2831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramachandran G.N., Ramakrishnan C., Sasisekharan V. Stereochemistry of polypeptide chain configurations. J. Mol. Biol. 1963;7:95–99. doi: 10.1016/s0022-2836(63)80023-6. [DOI] [PubMed] [Google Scholar]
- Renetseder R., Brunie S., Dijkstra B.W., Drenth J., Sigler P.B. A comparison of the crystal structures of phospholipase A2 from bovine pancreas and Crotalus atrox venom. J. Biol. Chem. 1985;260:11627–11634. [PubMed] [Google Scholar]
- Rimbault C., Maruthi K., Breillat C., Genuer C., Crrespillo S. Engineering selective competitors for the discrimination of highly conserved protein-protein interaction modules. Nat. Commun. 2019;10:4521. doi: 10.1038/s41467-019-12528-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rokyta D., Wray K.P., Lemmon A.R., Lemmon E.M., Caudle S.B. A high-throughput venom-gland transcriptome for the Eastern Diamondback Rattlesnake (Crotalus adamanteus) and evidence for pervasive positive selection across toxin classes. Toxicon. 2011;57:657–671. doi: 10.1016/j.toxicon.2011.01.008. [DOI] [PubMed] [Google Scholar]
- Rolland T., Tasan M., Charloteaux B., Pevzner S.J., Zhong Q., Sahni N., Yi S. A proteome-scale map of the human interactome network. Cell. 2014;159:1212–1226. doi: 10.1016/j.cell.2014.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ŝali A., Blundell T.L. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- Salvador G.H.M., Cardoso F.F., Gomes A.A., Cavalcante W.L.G., Gallacci M., Fontes M.R.M. Search for efficient inhibitors of myotoxic activity induced by ophidian phospholipase A2-like proteins using functional, structural and bioinformatics approaches. Sci. Rep. 2019;9:510. doi: 10.1038/s41598-018-36839-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador G.H.M., Dreyer T.R., Gomes A.A.S., Cavalcante W.L.G., Dos Santos J.I., Gandin C.A., de Oliveira Neto M., Gallacci M., Fontes M.R.M. Structural and functional characterization of suramin-bound MjTX-I from Bothrops moojeni suggests a particular myotoxic mechanism. Sci. Rep. 2018;8 doi: 10.1038/s41598-018-28584-7. 10317–10317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakano T., Mahamood M.I., Yamashita T., Fujitani H. Molecular dynamics analysis to evaluate docking pose prediction. Biophys. Physicobiol. 2016;13:181–194. doi: 10.2142/biophysico.13.0_181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shan Y., Kim E.T., Eastwood M.P., Dror R.O., Seeliger M.A., Shaw D.E. How does a drug molecule find its target binding site? J. Am. Chem. Soc. 2011;133:9181–9183. doi: 10.1021/ja202726y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiol N., Tadokoro T., Shioi S., Okabe Y., Matsubara H. Crystal structure of the complex between venom toxin and serum inhibitor from Viperidae snake. J. Biol. Chem. 2019;294:1250–1256. doi: 10.1074/jbc.RA118.006840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonis N., Rual J.F., Carvunis A.R., Tasan M., Lemmens I., Hirozane-Kishikawa T. Empirically controlled mapping of the Caenorhabditis elegans protein-protein interactome network. Nat. Methods. 2009;6:47–54. doi: 10.1038/nmeth.1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suryamohan K., Krishnankutty S.P., Guillory J., Jevit M. The Indian cobra reference genome and transcriptome enables comprehensive identification of venom toxins. Nat. Genet. 2020;52:106–117. doi: 10.1038/s41588-019-0559-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tadokoro T., Modahl C.M., Maenaka K., Aoki-Shioi N. Cysteine-rich secretory proteins (CRISPs) from venomous snakes: an overview of the functional diversity in a large and underappreciated superfamily. Toxins. 2020;12:174–175. doi: 10.3390/toxins12030175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Team R.C. R Foundation for Statistical Computing; Vienna, Austria: 2013. R: A Language and Environment for Statistical Computing.http://www.R-project.org/2013 [Google Scholar]
- Trigg S.A., Garza R.M., MacWilliams A., Nery J.R., Bartlett A., Castanon R., Goubil A., Feeney J., O’Malley R., Hung S.S.C., Zhang Z., Galli M., Ecker J.R. CrY2H-seq: a massively multiplexed assay for deep-coverage interactome mapping. Nat. Methods. 2017;14:819–825. doi: 10.1038/nmeth.4343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Zundert G.C.P., Rodrigues J.P.G.L.M., Trellet M., Schmitz C., Kastritis P.L., Karaca E., Melquiond A.S.J., van Dijk M., de Vries S.J., Bonvin A.M.J.J. The HADDOCK 2.2 webserver: user-friendly integrative modeling of biomolecular complexes. J. Mol. Biol. 2016;428:720–725. doi: 10.1016/j.jmb.2015.09.014. [DOI] [PubMed] [Google Scholar]
- Vangone A., Rodrigues J.P.G.L.M., Xue L.C., van Zundert G.C.P. Sense and simplicity in HADDOCK scoring: lessons from CASP-CAPRI round 1. Proteins. 2017;85:417–423. doi: 10.1002/prot.25198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webb B., Ŝali A. Comparative protein structure modeling using MODELLER. Curr. Protoc. Bioinform. 2014;47:5–6. doi: 10.1002/0471250953.bi0506s47. [DOI] [PubMed] [Google Scholar]
- WHO . 2018. Snakebite Envenomation Turns Again into a Neglected Tropical Disease! Available online at: https://www.who.int/snakebites/resources/s40409-017-0127-6/en/ (accessed 2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiederstein M., Sippl M.J. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res. 2007;35:W407–W410. doi: 10.1093/nar/gkm290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki Y., Morita T. Structure and function of snake venom cysteine-rich secretory proteins. Toxicon. 2004;44:227–231. doi: 10.1016/j.toxicon.2004.05.023. [DOI] [PubMed] [Google Scholar]
- Yu H., Braun P., Yildirim M.A., Lemmens I., Venkatesan K., Sahalie J. High-quality binary protein interaction map of the yeast interactome network. Science. 2008;322:104–110. doi: 10.1126/science.1158684. [DOI] [PMC free article] [PubMed] [Google Scholar]