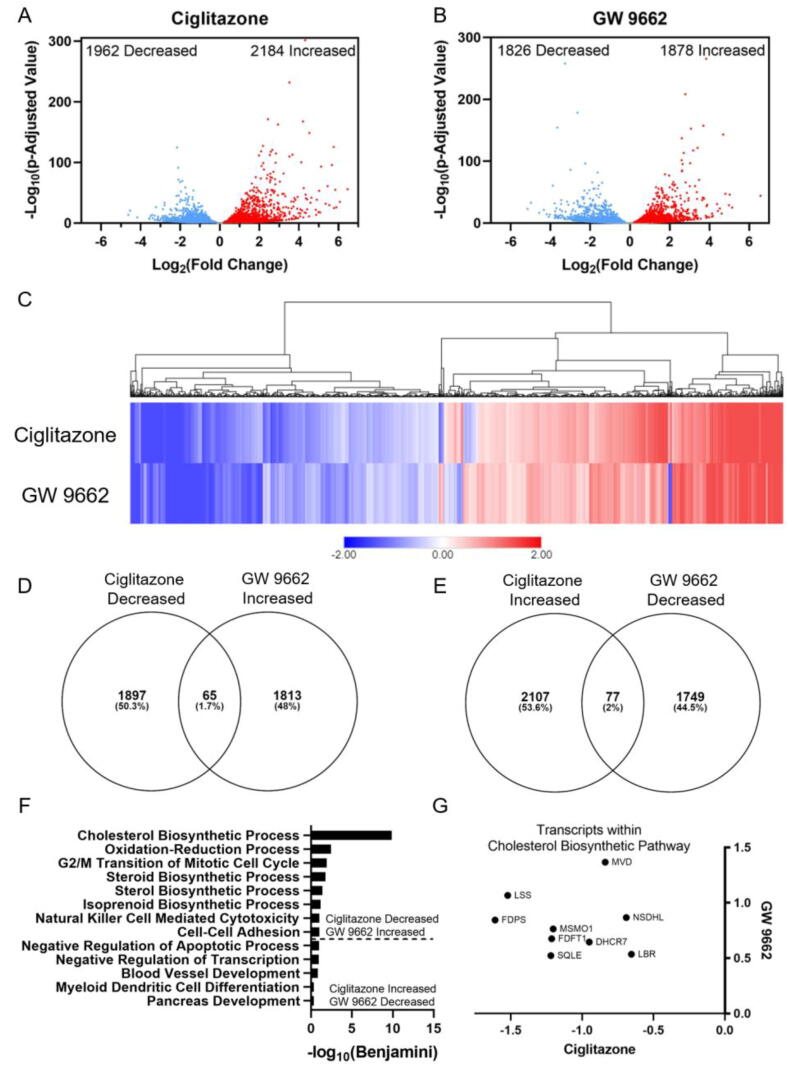
Fig. 5.
Volcano plots indicating number of significantly affected transcripts for ciglitazone (A) or GW 9662 (B) relative to vehicle-exposed cells. Heat map of significantly affected transcripts organized by hierarchical clustering using Euclidean distance and complete linkage method (C). Venn diagrams showing overlap in transcripts between ciglitazone and GW 9662 (D, E). Gene ontology analysis of biological processes identified by DAVID based on decreased transcripts following ciglitazone exposure and increased transcripts following GW 9662 exposure or increased transcripts following ciglitazone exposure and decreased transcripts following GW 9662 exposure (E). Transcripts within cholesterol biosynthetic process plotted by log2(fold change) in GW 9662-exposed cells along the Y-axis, and log2(fold change) in ciglitazone-exposed cells along the x-axis (F). MVD: Mevalonate Diphosphate Decarboxylase, NSDHL: Sterol-4-alpha-caroxylate 3-dehydrogenase (NAD(P) dependent steroid dehydrogenase-like), LBR: Lamin B receptor, DHCR7: 7-Dehydrocholesterol Reductase, MSMO1: Methylsterol Monooxygenase 1, FDFT1: Farnesyl-Diphosphate Farnesyltransferase 1, SQLE: Squalene Epoxidase, LSS: Lanosterol synthase, FDPS: Farnesyl pyrophosphate synthase.