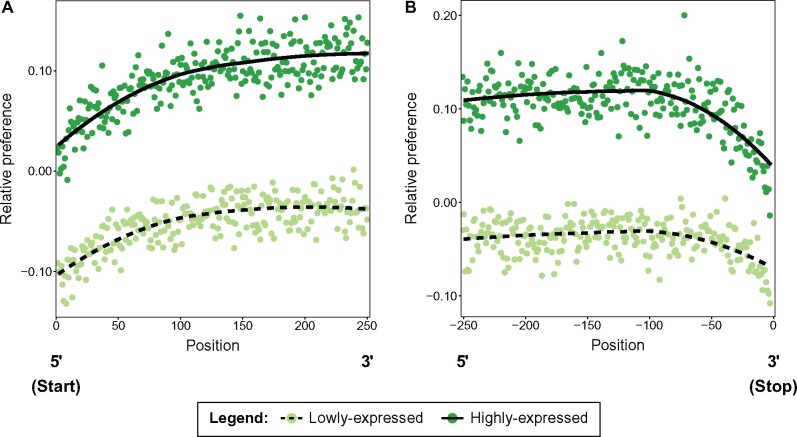
Fig. 6.
Patterns of codon preference across codon positions within lowly and highly expressed genes. In both plots, the lines represent splines (from an LOESS model; solid line = highly expressed genes, dashed line = lowly expressed genes) and the points represent the individual estimates at each codon position (darker points = highly expressed genes, lighter points = lowly expressed genes). (A) Pattern of codon preference starting from the beginning of genes, (B) pattern of codon preference leading up to the stop codon (indicated by position zero, so negative positions are distances before the stop codon). Average preference of codons is measured relative to (i.e., as a deviation from) the genome-wide value.