Fig. 7.
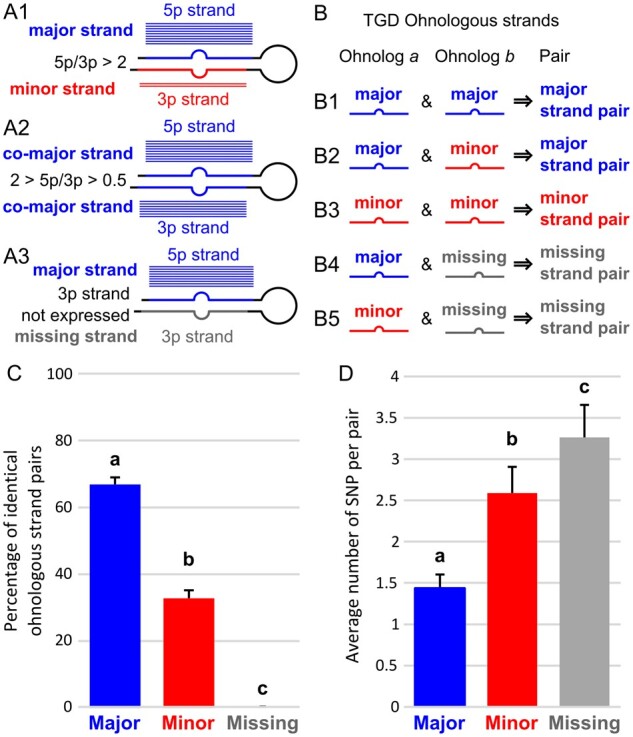
Global pattern of sequence evolution between ohnologous mature miRNAs. (A) Each strand in a hairpin was qualified as either major or minor depending on its relative expression compared with the complementary strand of the hairpin. Blue and red horizontal bars illustrate relative expression patterns (in analogy to aligned reads). Strands that were not expressed were qualified as missing. (B) Ohnologous mature miRNA pairs were further qualified as major strand pairs, minor strand pairs, and missing strand pairs depending on the qualification of each of the mature miRNA of the ohnologous pair. (C) Percentage of ohnologous miRNA strand pairs that were composed of identical miRNAs. Values represent teleost averages with SD for major, minor, and missing strand pairs. (D) Average number of single-nucleotide polymorphisms (SNPs) in ohnologous strand pairs that were not identical. Values represent teleost averages with SDs for major, minor, and missing strand pairs. Different letters signify significant differences at P < 0.05.
