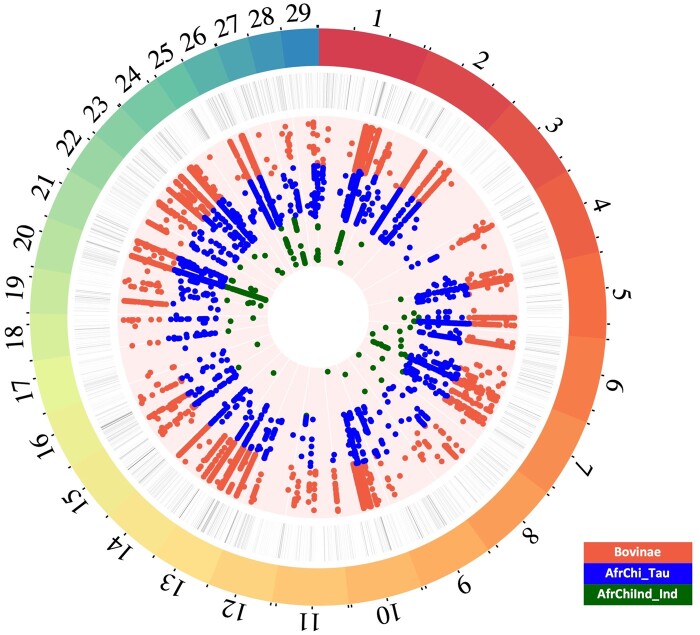
Fig. 4.
High frequency SNPs in Yakut cattle but low frequency/absence of the same allele in Holstein, Kholmogory, and Hanwoo (total of 13,257 SNP). Each circle from the periphery to the center shows the following data sets: bovine autosomes, indicine-like genome intervals from the RFMix analysis in different shades of gray (light to dark indicating indicine-like interval frequencies in the Yakut population ranging from 50% to 100%), Yakut alleles found in Bovinae species in red, in African and Chinese taurine breeds in blue, and exclusively in Indian, African, and Chinese indicine breeds in green.