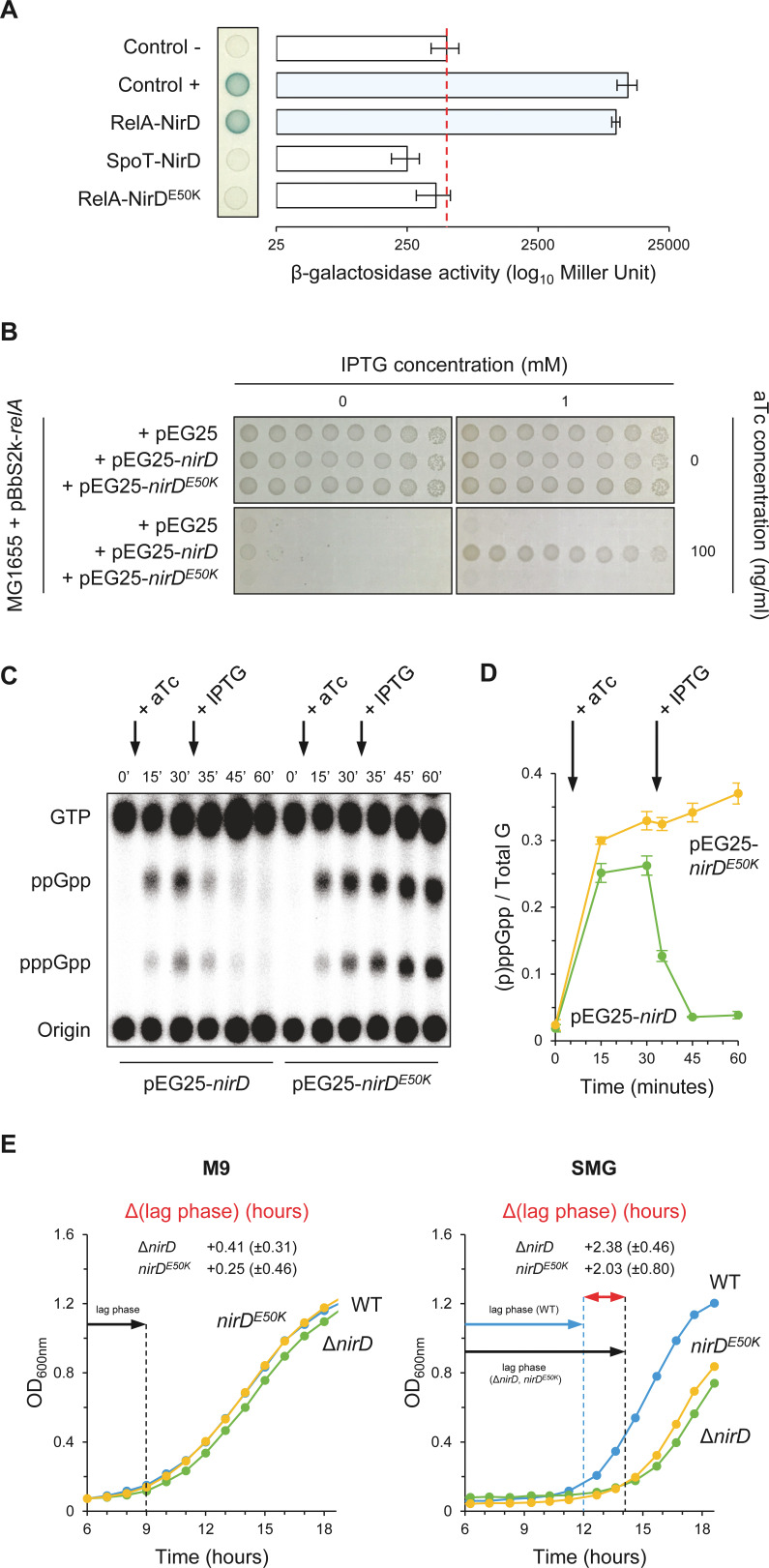
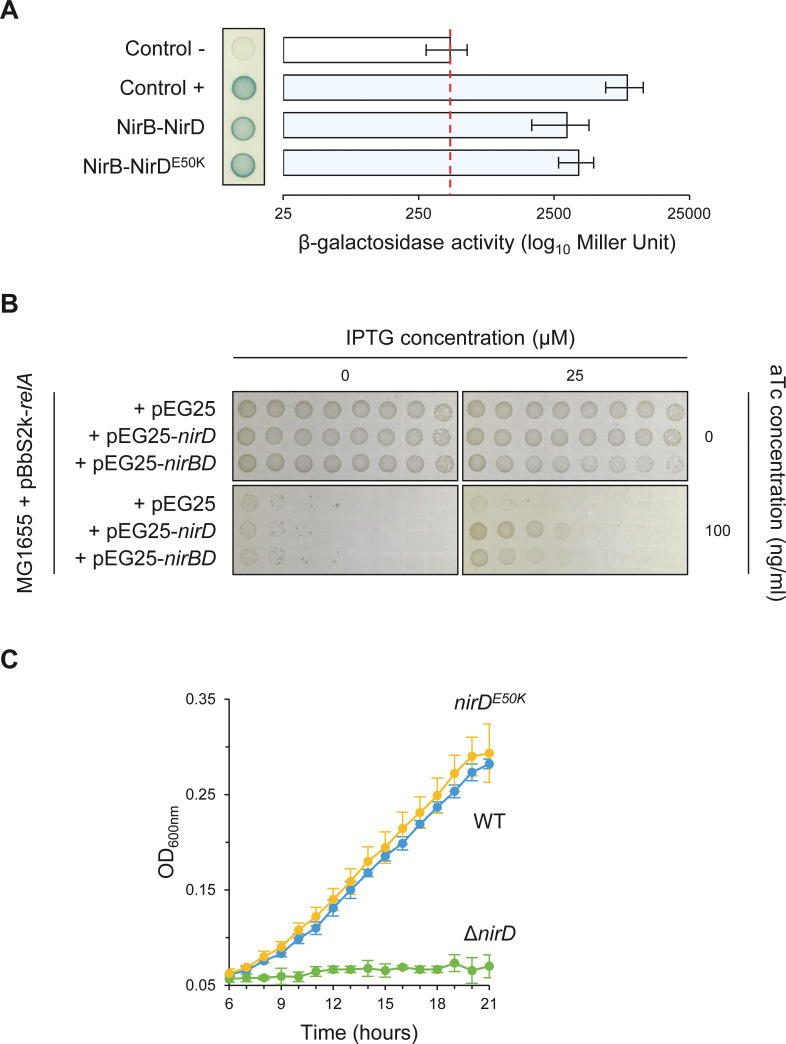
Figure 3. NirD can inhibit RelA-dependent accumulation through physical interaction in vivo.
(A) Bacterial two-hybrid assay using E. coli BTH101 cells co-transformed with plasmid derivatives pKT25-relA or pKT25-spoT and pUT18C-nirD or pUT18C-nirDE50K. Stationary-phase cultures were spotted on NA plates containing X-Gal as a blue color reporter for positive interaction. The bars showing β-galactosidase activity are represented as the means of three independent experiments, and the error bars depict the SDs. (B) Growth assay of E. coli MG1655 cells co-transformed with the plasmid derivatives pBbS2k-relA and pEG25-nirD or pEG25-nirDE50K inducible by anhydrotetracycline (aTc) and isopropyl β-D-1-thiogalactopyranoside (IPTG), respectively. Serial dilutions of stationary-phase cultures were spotted on NA plates containing the indicated concentrations of inducers. The results are representative of three independent experiments. (C, D) In vivo (p)ppGpp level dynamic in E. coli MG1655 cells carrying pBbS2k-relA and pEG25-nirD or pEG25-nirDE50K after successive inductions of expression of the relA and nirD or nirDE50K genes using 100 ng/mL aTc and 1 mM IPTG, respectively. Nucleotides extracted from samples collected at the indicated times were separated by thin layer chromatography. The autoradiogram (C) is representative of three independent experiments, and the curves of the relative levels of (p)ppGpp (D) are represented as the means of the three independent experiments, the error bars depicting the SDs. (E) Growth curves of wild-type (WT) cells, ΔnirD, and nirDE50K mutants under anaerobiosis in M9-glucose minimal medium without amino acids (left panel) or supplemented with 1 mM serine, methionine, and glycine (SMG) (right panel) (see Materials and methods). The growth curves are representative of at least three independent experiments, and the Δ(lag phase) are represented as the means (± SD) of the independent experiments.