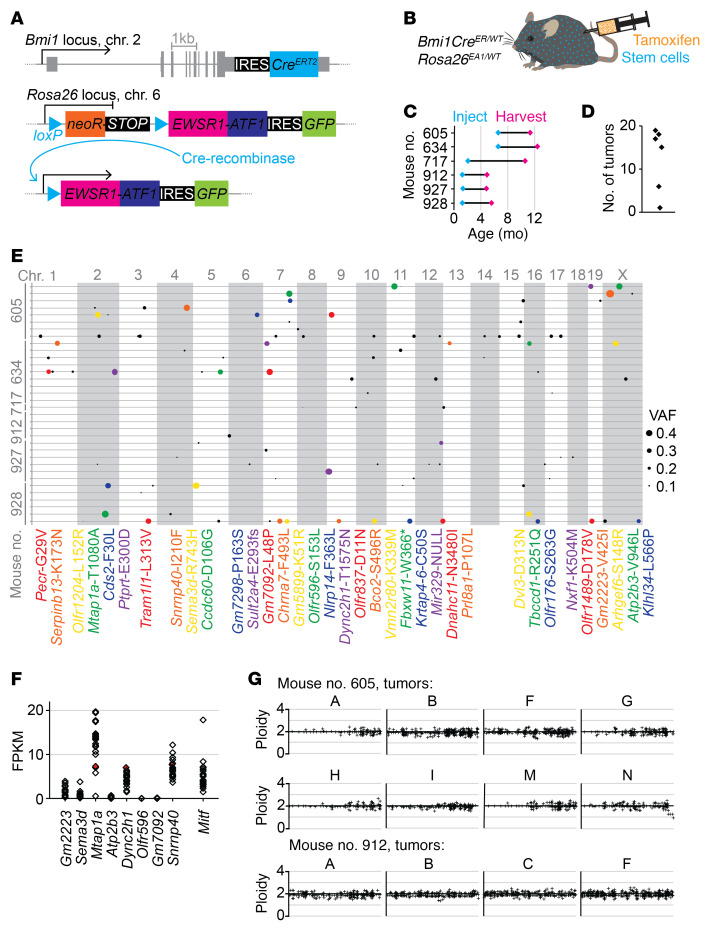
Figure 3. Exome sequencing of EWSR1-ATF1 expression–initiated mouse tumors reveals that no secondary alterations are strictly required to complete sarcomagenesis.
(A) Schematic illustrating CreERT2 engineered at the Bmi1 locus in conjunction with Cre-inducible human EWSR1-ATF1 engineered at the Rosa26 locus. Cre recombinase removes the neoR-STOP cassette to induce expression of EWSR1-ATF1, and the GFP reporter confirms EWSR1-ATF1 expression. (B) Tamoxifen injection induces CreERT2 activity in Bmi1-expressing stem cells (blue), and then in turn, Cre recombinase induces Rosa26-mediated expression of EWSR1-ATF1 (EA1). (C) Time course showing age at injection and age at tumor harvesting (n = 6). (D) Summary of total number of tumors detected and harvested per mouse (n = 6). (E) Exome sequencing of 34 mouse tumors, with 1 tumor’s exome presented on each line, clustered by host mouse, and each variant allele denoted by a circle whose size corresponds to the VAF. All variants with a VAF of greater than 0.25 are also identified by corresponding colors in the list of gene symbols and protein amino acid substitutions below. (F) RNA-Seq rendered expression levels relative to Gapdh for the 8 genes with variants present at fractions higher than 0.4. The level of Mitf expression is included for reference. (G) Exome-wide CNV was inferred from exome sequencing data, as shown for 12 mouse tumors. Normal diploidy is represented by the x axis crossing at a value of y = 2.