Figure 1. Identification of protein kinase D (PKD)3 substrates by immunoprecipitation using PKD substrate motif antibody LxRxx[S*/T*].
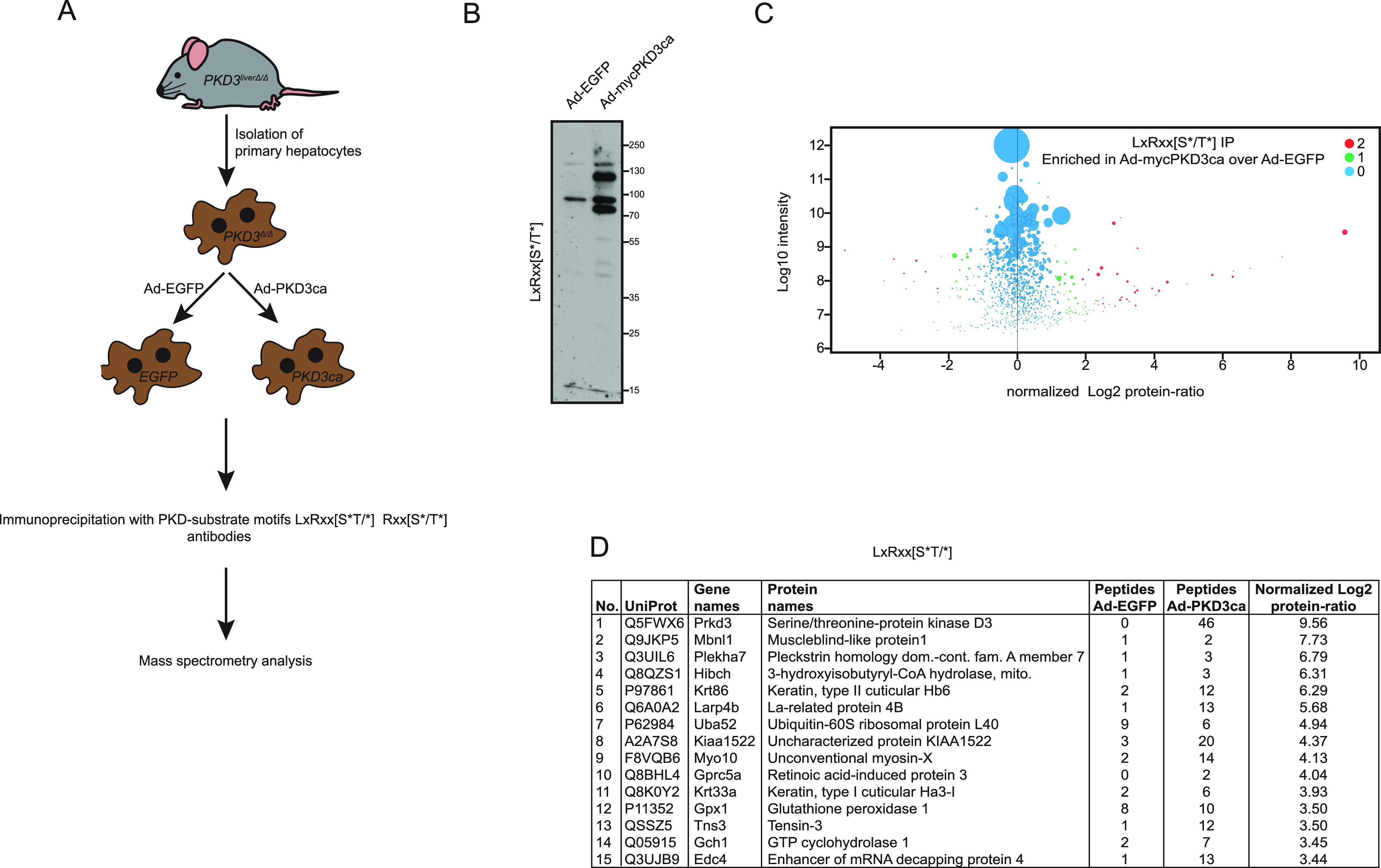
(A) Experimental design. Primary hepatocytes isolated from three PKD3-deficient mice were transduced by adenovirus containing EGFP (controls) or the constitutive active form of PKD3 (PKD3ca). IP with PKD-substrate motif antibodies was performed on cell extracts and followed by mass spectrometry analysis. (B) WB analysis of protein lysates from PKD3-deficient primary hepatocytes transduced with either adenovirus expressing control EGFP (Ad-EGFP) or constitutive active PKD3 (Ad-mycPKD3ca) using PKD-substrate motif LxRxx[S*/T*]-specific antibody (n = 3 independent experiments). (C) Scatter plot of the statistical significance of log2 transformed protein ratios versus log10-transformed label-free quantification intensities between control and PKD3ca expressing hepatocytes. Enriched proteins are indicated by red (2, significantly enriched) or green (1, potentially enriched) dots, blue ones are not enriched (0, not enriched). (D) 15 most enriched proteins identified by mass spectrometry showing a number, UniProt accession number, gene names, protein names, peptide count in EGFP and PKD3ca samples, respectively, and log2-transformed label-free quantification protein ratio are shown in the table. The detailed list of proteins identified are listed in Table S1.
