Figure 3. Comparative analysis of putative protein kinase D (PKD)3 targets identified using different motif antibodies.
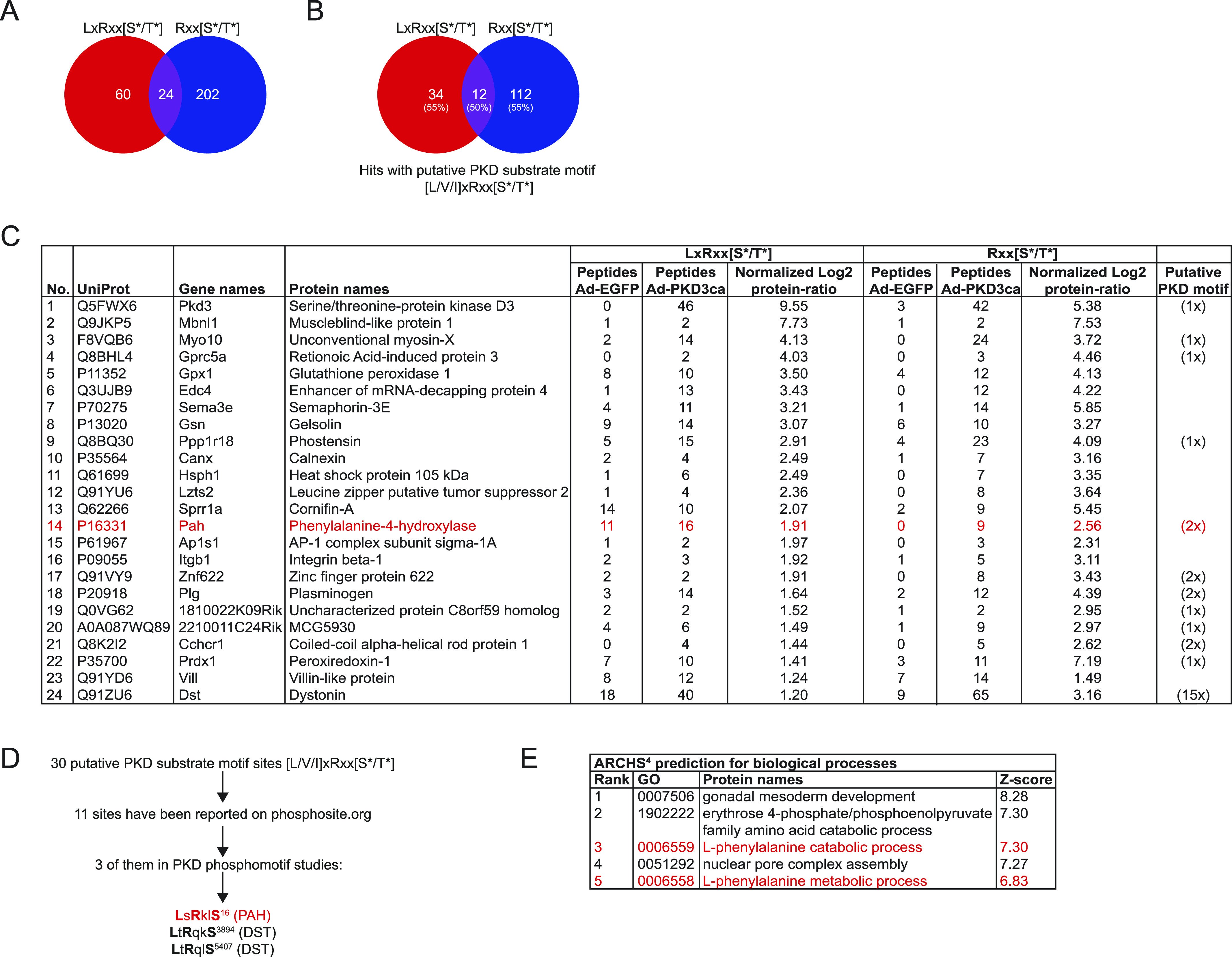
(A) Venn diagram of common putative substrates that were significantly enriched by both antibodies LxRxx[S*/T*] and Rxx[S*/T*]. (B) Computational identification of putative PKD motifs [L/V/I]xRxx[S*/T*] among putative substrates identified by both antibodies (percentage of proteins possessing a putative motif in brackets). Results were obtained using ExPASy ScanProsite tool. (C) List of 24 proteins enriched by both antibodies, LxRxx[S*/T*] and Rxx[S*/T*] with UniProt accession numbers, gene and protein names, peptide count, and log2 transformed label-free quantification protein ratio for LxRxx[S*/T*] and Rxx[S*/T*] immunoprecipitations, respectively, and the amount of putative PKD motifs for each protein. (D) Flow of computational identification of putative PKD substrates by comparing the PKD motifs within proteins using phosphosite.org repository. (E) Prediction of biological processes potentially regulated by PKD3 using ARCHS4.
