Figure 4. IL-13–dependent lung proteomic changes during Nippostrongylus brasiliensis infection.
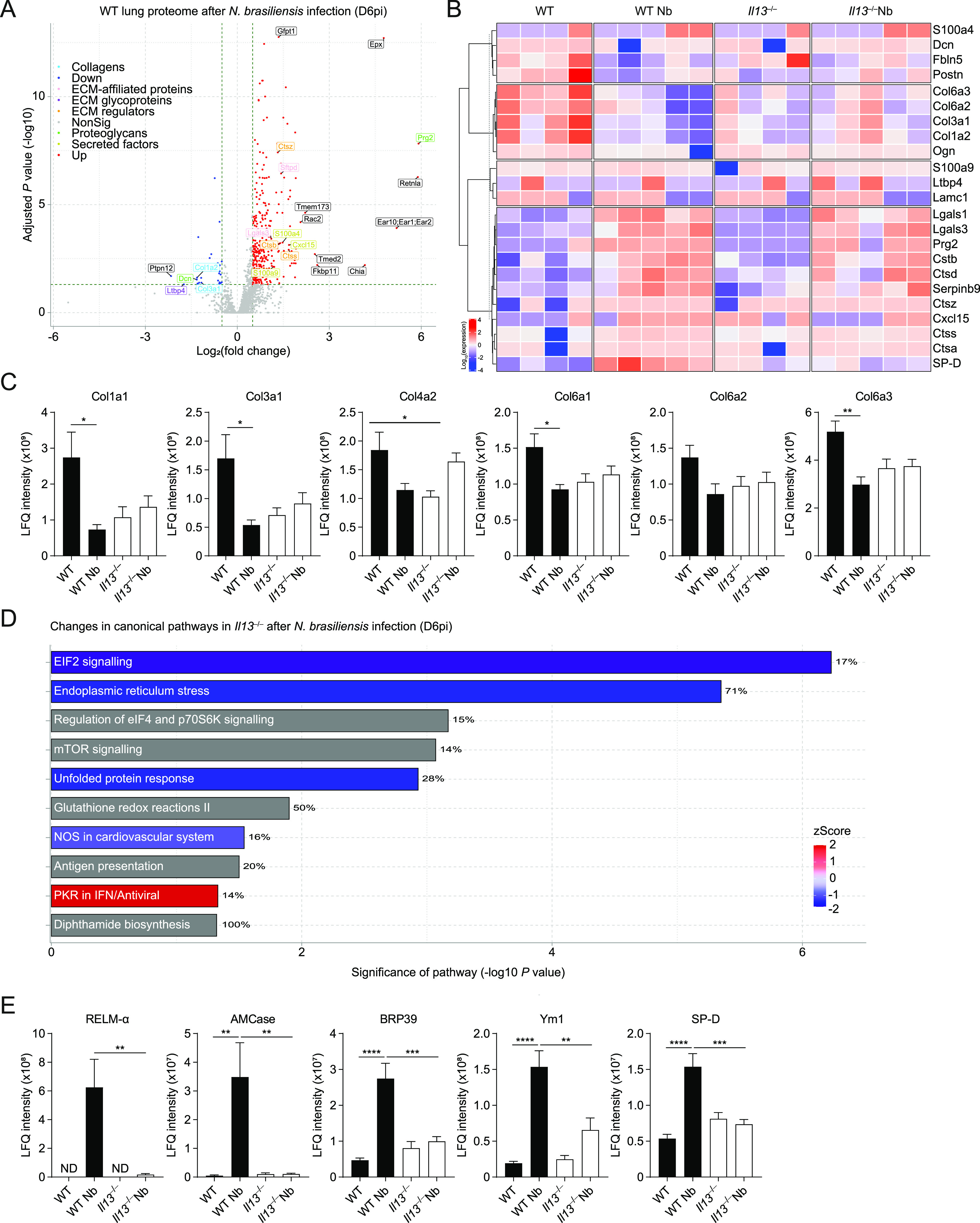
WT and Il13−/− mice were infected with N. brasiliensis (Nb) and on D6pi lungs were prepared for proteomic analysis. (A) Volcano plot of infected WT lungs (D6pi) showing differential expression of up- (red) or down- (blue) regulated proteins with matrisome annotations (fold change relative to naïve WT mice). Black labels indicate proteins lacking matrisome annotations. (B) Unsupervised, hierarchically clustered heatmap of expression of matrisome proteins comparing naïve and Nb-infected groups of WT and Il13−/− mice on D6pi. (C) Columns in each set represent different (biological repeat) mice in each group (C) Relative abundance (label-free quantification [LFQ] intensity) of collagen peptides comparing infected WT and Il13−/− lungs on D6pi. (D) Predicted changes in canonical pathways based on changes in the proteome of infected Il13−/− mice compared with infected WT mice (down-regulation in blue, up-regulation in red, and no specified direction in grey). Percentage indicates the relative number of proteins that are regulated in each pathway. (E) Relative abundance of peptides highly associated with type 2 immunity comparing infected WT and Il13−/− lungs on D6pi. Data (mean ± SEM in C and E) are pooled from two independent mass spectrometry runs with four to five mice per group in total. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (one-way ANOVA and Tukey–Kramer post hoc test).
