Figure 6. Transcriptional profiling of IL-13–dependent genes in the lung after Nippostrongylus brasiliensis infection.
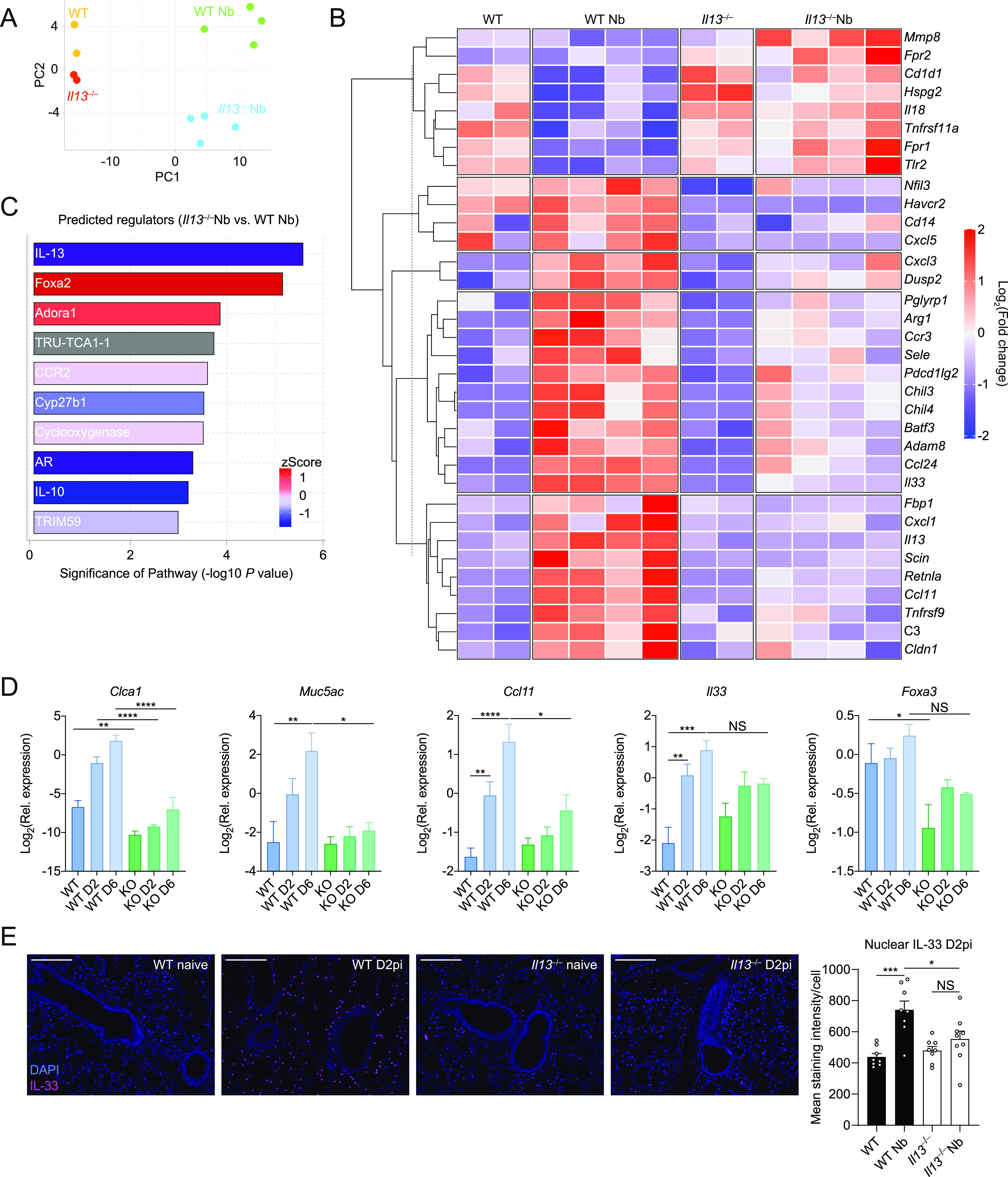
Whole lung RNA from WT and Il13−/− mice infected with N. brasiliensis (Nb) on D6pi was analysed by Nanostring. (A) Principle components analysis of naïve and infected WT and Il13−/− mice. (B) Unsupervised, hierarchically clustered heat map of genes differentially expressed between mouse groups with fold change expression level indicated by colour. (C) Columns in each set represent different (biological repeat) mice in each group (C) Predicted upstream regulators from Ingenuity Pathway Analysis. (D) Expression of Foxa2-regulated genes Clca1, Muc5ac, Ccl11, Il33, and Foxa3 were measured in lung tissues on day 2 post-infection and D6pi by quantitative real-time PCR (data normalised against housekeeping gene Rpl13a). (E) Immunofluorescence staining of nuclear IL-33 (magenta) in the parenchyma of the lung and quantification of mean integrated density (IntDen) (scale bar = 100 µm). Data in (A, B, C) are from a single Nanostring run with samples from two to four mice per group. Data (mean ± SEM) in (D, E) were pooled from two individual experiments with three to five mice per group (per experiment). NS, not significant, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001 (one-way ANOVA and Tukey–Kramer post hoc test).
